5F3T
 
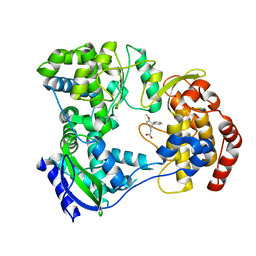 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to JF-31-MG46 | | Descriptor: | 2-(4-methoxy-3-phenyl-phenyl)ethanoic acid, RNA-DEPENDENT RNA POLYMERASE, ZINC ION | | Authors: | Noble, C.G. | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Conserved Pocket in the Dengue Virus Polymerase Identified through Fragment-based Screening.
J.Biol.Chem., 291, 2016
|
|
3O2Q
 
 | |
7PRD
 
 | |
8RHS
 
 | |
1FOX
 
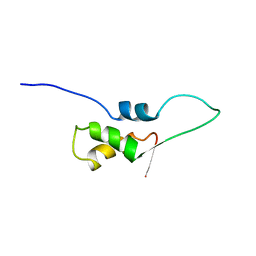 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURES | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
8B07
 
 | |
1FOW
 
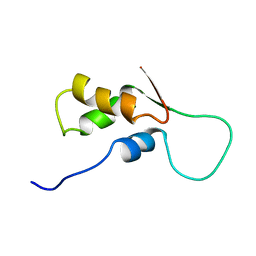 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
4X9C
 
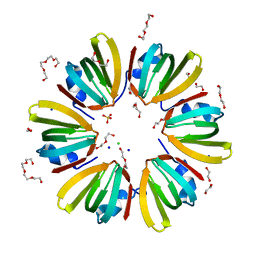 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
4X9D
 
 | | High-resolution structure of Hfq from Methanococcus jannaschii in complex with UMP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, E.Y, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
8G9O
 
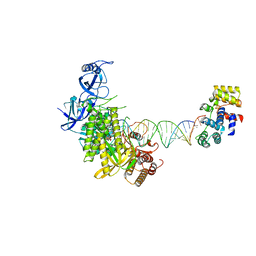 | | Complete DNA elongation subcomplex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HKV
 
 | |
6GVV
 
 | |
8CEV
 
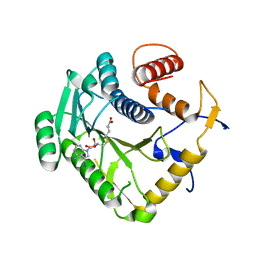 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO1119 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-iodanyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CER
 
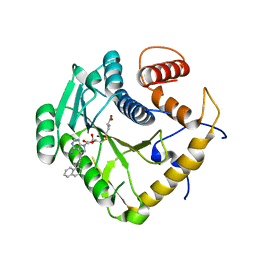 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO494 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-naphthalen-1-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CEQ
 
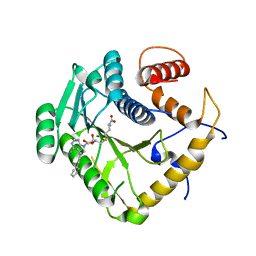 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO427 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-phenylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CES
 
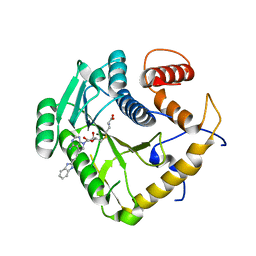 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO500 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-[2-(1~{H}-benzimidazol-2-yl)ethynyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
8CET
 
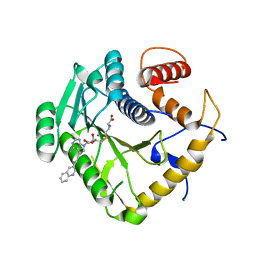 | | Crystal structure of monkeypox virus methyltransferase VP39 in complex with inhibitor TO507 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase | | Authors: | Klima, M, Silhan, J, Boura, E. | | Deposit date: | 2023-02-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
3ZZS
 
 | | Engineered 12-subunit Bacillus stearothermophilus trp RNA-binding attenuation protein (TRAP) | | Descriptor: | TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Chen, C, Smits, C, Dodson, G.G, Shevtsov, M.B, Merlino, N, Gollnick, P, Antson, A.A. | | Deposit date: | 2011-09-02 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | How to Change the Oligomeric State of a Circular Protein Assembly: Switch from 11-Subunit to 12-Subunit Trap Suggests a General Mechanism
Plos One, 6, 2011
|
|
8CGB
 
 | | Monkeypox virus VP39 in complex with SAH | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Silhan, J, Klima, M, Skvara, P, Boura, E. | | Deposit date: | 2023-02-03 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase.
Nat Commun, 14, 2023
|
|
5A2V
 
 | | Crystal structure of mtPAP in Apo form | | Descriptor: | CHLORIDE ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A30
 
 | | Crystal structure of mtPAP N472D mutant in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOCHONDRIAL PROTEIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2Z
 
 | | Crystal structure of mtPAP in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
5A2X
 
 | | Crystal structure of mtPAP in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
8INB
 
 | | Cryo-EM structure of Cas12j-SF05-crRNA-dsDNA complex | | Descriptor: | Cas12j-SF05, NTS-DNA, TS-DNA, ... | | Authors: | Zhang, X, Duan, Z.Q, Zhu, J.K. | | Deposit date: | 2023-03-09 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for DNA cleavage by the hypercompact Cas12j-SF05.
Cell Discov, 9, 2023
|
|
6IFR
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-NTR, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Type III-A CRISPR-associated RAMP protein Csm3, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
