8U6M
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(2-((6-chloro-2-cyanoindolizin-8-yl)oxy)phenoxy)ethyl)-N-methylacrylamide (JLJ751), a non-nucleoside inhibitor | | Descriptor: | N-[2-(2-{[(4R)-6-chloro-2-cyanoindolizin-8-yl]oxy}phenoxy)ethyl]-N-methylpropanamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
5H5B
 
 | |
5TUJ
 
 | |
8U6D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with N-(2-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenoxy)ethyl)-N-methylacrylamide (JLJ736), a non-nucleoside inhibitor | | Descriptor: | N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenoxy]ethyl}-N-methylprop-2-enamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
7OZC
 
 | |
5C5W
 
 | | 1.25 A resolution structure of an RNA 20-mer | | Descriptor: | RNA (5'-R(P*CP*CP*UP*GP*AP*GP*UP*UP*CP*AP*AP*UP*UP*CP*UP*AP*GP*CP*G)-3') | | Authors: | Stewart, M, Valkov, E. | | Deposit date: | 2015-06-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 angstrom resolution structure of an RNA 20-mer that binds to the TREX2 complex.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8U6S
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 8-(2-(3-morpholino-3-oxopropoxy)phenoxy)indolizine-2-carbonitrile (JLJ757), a non-nucleoside inhibitor | | Descriptor: | (4S)-8-{2-[3-(morpholin-4-yl)-3-oxopropoxy]phenoxy}indolizine-2-carbonitrile, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
7OQW
 
 | |
7ORH
 
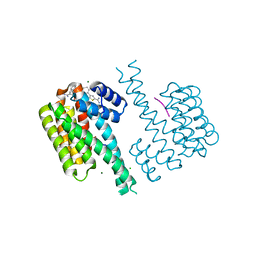 | | Ternary complex of 14-3-3 sigma, p27pT198 phosphopeptide, and WQ178 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Centorrino, F, Wu, Q, Ottmann, C. | | Deposit date: | 2021-06-05 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A crystallography-based study of fragment extensions
into the 14-3-3 binding groove
To Be Published
|
|
8U6C
 
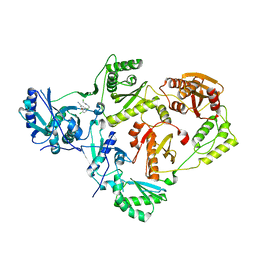 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 2-chloro-N-(4-chloro-3-(3-chloro-5-cyanophenoxy)phenethyl)acetamide (JLJ732), a non-nucleoside inhibitor | | Descriptor: | 2-chloro-N-{2-[4-chloro-3-(3-chloro-5-cyanophenoxy)phenyl]ethyl}acetamide, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Hollander, K, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8JV3
 
 | |
7XUV
 
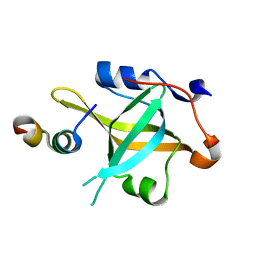 | | Crystal structure of RPA70N-RMI1 fusion | | Descriptor: | RecQ-mediated genome instability protein 1, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
7OZ8
 
 | |
7MKV
 
 | |
6MGQ
 
 | | ERAP1 in the open conformation bound to 10mer phosphinic inhibitor DG014 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, Phosphinic inhibitor DG014, ... | | Authors: | Stern, L.J, Maben, Z. | | Deposit date: | 2018-09-14 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Conformational dynamics linked to domain closure and substrate binding explain the ERAP1 allosteric regulation mechanism.
Nat Commun, 12, 2021
|
|
7OXN
 
 | | Crystal Structure of TAP01 in complex with cyclised amyloid beta peptide | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta precursor protein, TAP01 family antibody heavy chain, ... | | Authors: | Hall, G, Cowan, R, Carr, M.D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a novel pseudo beta-hairpin structure of N-truncated amyloid-beta for use as a vaccine against Alzheimer's disease.
Mol Psychiatry, 27, 2022
|
|
8QNL
 
 | |
8U6R
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-((2-cyanoindolizin-8-yl)oxy)phenoxy)-N-(2,2-difluoroethyl)propanamide (JLJ756), a non-nucleoside inhibitor | | Descriptor: | 3-(2-{[(4R)-2-cyanoindolizin-8-yl]oxy}phenoxy)-N-(2,2-difluoroethyl)propanamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Henry, S, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
8UPI
 
 | | Structure of a periplasmic peptide binding protein from Mesorhizobium sp. AP09 bound to aminoserine | | Descriptor: | 1,2-ETHANEDIOL, AMINOSERINE, CALCIUM ION, ... | | Authors: | Frkic, R.L, Smith, O.B, Rahman, M, Kaczmarski, J.A, Jackson, C.J. | | Deposit date: | 2023-10-22 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Characterization of a Bacterial Periplasmic Solute Binding Protein That Binds l-Amino Acid Amides.
Biochemistry, 63, 2024
|
|
8JWV
 
 | | Untethered R0RBR | | Descriptor: | BARIUM ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
7MCI
 
 | | MoFe protein from Azotobacter vinelandii with a sulfur-replenished cofactor | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Lee, C, Hu, Y, Ribbe, M.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor
Nat Catal, 5, 2022
|
|
7P26
 
 | |
5UBH
 
 | | Catalytic core domain of Adenosine triphosphate phosphoribosyltransferase from Campylobacter jejuni with bound ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Mittelstaedt, G, Jiao, W, Livingstone, E.K, Parker, E.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A dimeric catalytic core relates the short and long forms of ATP-phosphoribosyltransferase.
Biochem. J., 475, 2018
|
|
8JXK
 
 | | Crystal Structure of Rv0047c from Mycobacterium tuberculosis | | Descriptor: | Conserved protein | | Authors: | Ansari, M.S, Yadav, V, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of Rv0047c from Mycobacterium tuberculosis
To Be Published
|
|
8UVZ
 
 | |
