1RDP
 
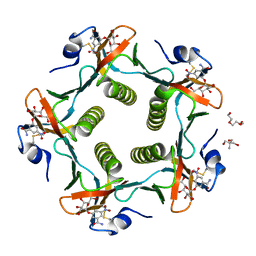 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV3 | | Descriptor: | 1,3-BIS-([[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBU TENYL]-AMINO-ETHYL]-AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RQ2
 
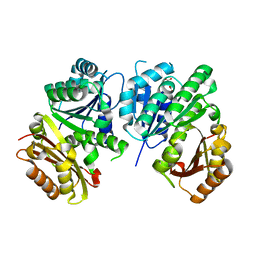 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH CITRATE | | Descriptor: | CITRIC ACID, Cell division protein ftsZ | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
1R1P
 
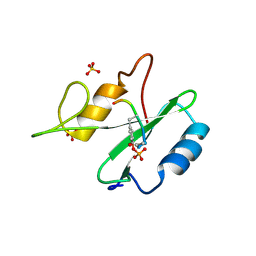 | |
1R6A
 
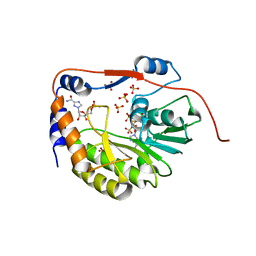 | | Structure of the dengue virus 2'O methyltransferase in complex with s-adenosyl homocysteine and ribavirin 5' triphosphate | | Descriptor: | Genome polyprotein, RIBAVIRIN MONOPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Benarroch, D, Egloff, M.P, Mulard, L, Romette, J.L, Canard, B. | | Deposit date: | 2003-10-15 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate.
J.Biol.Chem., 279, 2004
|
|
1RLU
 
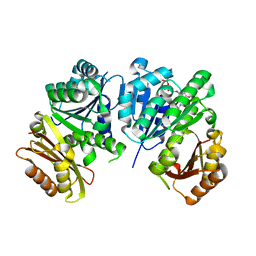 | | Mycobacterium tuberculosis FtsZ in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein ftsZ, GLYCEROL | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-11-26 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
4MFF
 
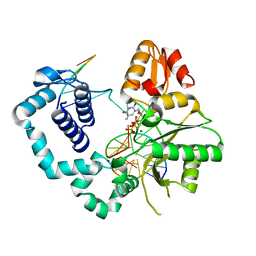 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable TTP | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
1R29
 
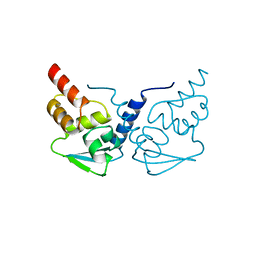 | | Crystal Structure of the B-Cell Lymphoma 6 (BCL6) BTB Domain to 1.3 Angstrom | | Descriptor: | B-cell lymphoma 6 protein | | Authors: | Ahmad, K.F, Melnick, A, Lax, S.A, Bouchard, D, Liu, J, Kiang, C.L, Mayer, S, Licht, J.D, Prive, G.G. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain.
Mol.Cell, 12, 2003
|
|
1R38
 
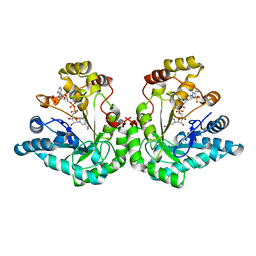 | | Crystal structure of H114A mutant of Candida tenuis xylose reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | Authors: | Kratzer, R, Kavanagh, K.L, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2003-09-30 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies of the enzymic mechanism of Candida tenuis xylose reductase (AKR 2B5): X-ray structure and catalytic reaction profile for the H113A mutant
Biochemistry, 43, 2004
|
|
1R94
 
 | |
4MFC
 
 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable CTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
1RVG
 
 | |
4HNA
 
 | |
1RH8
 
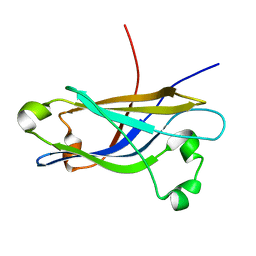 | | Three-dimensional structure of the calcium-free Piccolo C2A-domain | | Descriptor: | Piccolo protein | | Authors: | Garcia, J, Gerber, S.H, Sugita, S, Sudhof, T.C, Rizo, J. | | Deposit date: | 2003-11-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the Piccolo C2A domain regulated by alternative splicing.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3K45
 
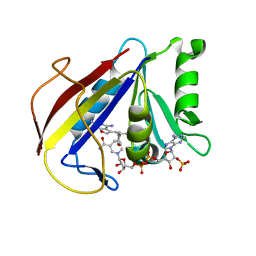 | | Alternate Binding Modes Observed for the E- and Z-isomers of 2,4-Diaminofuro[2,3d]pyrimidines as Ternary Complexes with NADPH and Mouse Dihydrofolate Reductase | | Descriptor: | 5-[(1Z)-2-(2-methoxyphenyl)prop-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, synthesis, and X-ray crystal structures of 2,4-diaminofuro[2,3-d]pyrimidines as multireceptor tyrosine kinase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
3RC7
 
 | |
3J2A
 
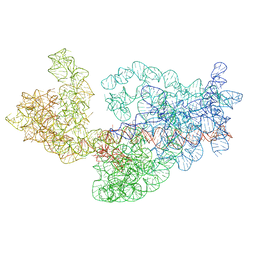 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.1 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
1TYD
 
 | |
5D22
 
 | |
6QC8
 
 | | Ovine respiratory complex I FRC open class 2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6QC7
 
 | | Ovine respiratory complex I FRC open class 3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6QBX
 
 | | Ovine respiratory supercomplex I+III2 closed class. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-24 | | Release date: | 2019-08-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
4D8C
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BXD552, derived from a co-crystallization experiment | | Descriptor: | (3S,4S,5R)-3-(4-amino-3-{[(2R)-3-ethoxy-1,1,1-trifluoropropan-2-yl]oxy}-5-fluorobenzyl)-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1, SULFATE ION | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
6QC4
 
 | | Ovine respiratory supercomplex I+III2 open class 3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Acyl carrier protein, Cytochrome b, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6QC5
 
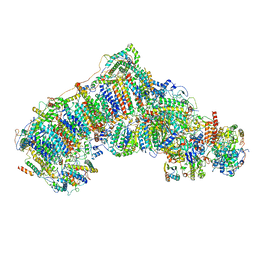 | | Ovine respiratory complex I FRC closed class 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Acyl carrier protein, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6QCF
 
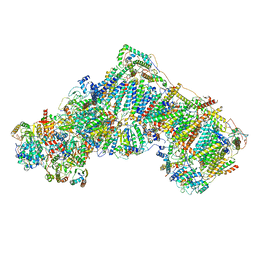 | | Ovine respiratory complex I FRC open class 6 | | Descriptor: | Acyl carrier protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
