2M1N
 
 | |
2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2M4J
 
 | | 40-residue beta-amyloid fibril derived from Alzheimer's disease brain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Lu, J, Qiang, W, Meredith, S.C, Yau, W, Schweiters, C.D, Tycko, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of beta-Amyloid Fibrils in Alzheimer's Disease Brain Tissue.
Cell(Cambridge,Mass.), 154, 2013
|
|
2LXU
 
 | | Solution NMR Structure of the eukaryotic RNA recognition motif, RRM1, from the heterogeneous nuclear ribonucleoprotein H from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8614A | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Ramelot, T.A, Yang, Y, Pederson, K, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the eukaryotic RNA recognition motif, RRM1, from the heterogeneous nuclear ribonucleoprotein H from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8614A
To be Published
|
|
2M52
 
 | |
2M7O
 
 | |
2M7S
 
 | | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1 | | Descriptor: | Serine/arginine-rich splicing factor 1 | | Authors: | Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for T-Cell Biology (TCELL) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of RNA recognition motif 2 (RRM2) of Homo sapiens splicing factor, arginine/serine-rich 1
To be Published
|
|
2MCQ
 
 | | NMR structure of a BolA-like hypothetical protein RP812 from Rickettsia prowazekii, Seattle structural genomics center for infectious disease (SSGCID) | | Descriptor: | Uncharacterized protein RP812 | | Authors: | Chen, Y, Barnwal, R, Yang, F, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-08-22 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a BolA-like hypothetical protein RP812 from Rickettsia prowazekii, Seattle structural genomics center for infectious disease (SSGCID)
To be Published
|
|
2M1E
 
 | |
2MLE
 
 | | NMR structure of the C-domain of troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Baryshnikova, O.K, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
2MN2
 
 | | 3D structure of YmoB, a modulator of biofilm formation | | Descriptor: | YmoB | | Authors: | Marimon, O, Cordeiro, T.N, Amata, I, Pons, M. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An oxygen-sensitive toxin-antitoxin system.
Nat Commun, 7, 2016
|
|
2M07
 
 | | NMR structure of OmpX in DPC micelles | | Descriptor: | Outer membrane protein X | | Authors: | Hagn, F.X, Etzkorn, M, Raschle, T, Wagner, G, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2012-10-21 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2MJC
 
 | | Zn-binding domain of eukaryotic translation initiation factor 3, subunit G | | Descriptor: | Eukaryotic translation initiation factor 3 subunit G, ZINC ION | | Authors: | Al-Abdul-Wahid, M, Menade, M, Xie, J, Kozlov, G, Gehring, K. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Zn-binding domain of eukaryotic translation initiation factor 3, subunit G
To be Published
|
|
2MMB
 
 | |
2M5L
 
 | |
2MLK
 
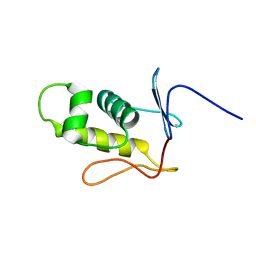 | | Three-dimensional structure of the C-terminal DNA-binding domain of RstA protein from Klebsiella pneumoniae | | Descriptor: | RstA | | Authors: | Fang, P, Chen, S, Cheng, Y, Chang, C, Yu, T, Huang, T. | | Deposit date: | 2014-03-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
4UX1
 
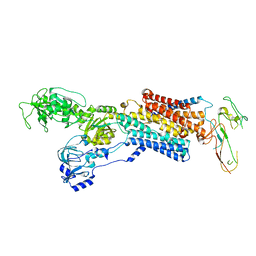 | | Cryo-EM structure of antagonist-bound E2P gastric H,K-ATPase (SCH.E2. AlF) | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Fujiyoshi, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-11-12 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Systematic Comparison of Molecular Conformations of H+,K+-ATPase Reveals an Important Contribution of the A-M2 Linker for the Luminal Gating.
J.Biol.Chem., 289, 2014
|
|
8XGA
 
 | |
6WDY
 
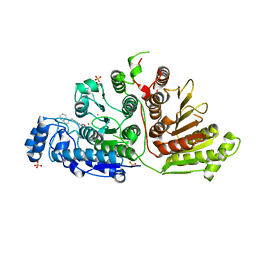 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with Indole Phenylhydroxamate Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-hydroxy-4-[(1H-indol-1-yl)methyl]benzamide, PHOSPHATE ION, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for the Selective Inhibition of HDAC10, the Cytosolic Polyamine Deacetylase.
Acs Chem.Biol., 15, 2020
|
|
4JZW
 
 | |
6WIC
 
 | | Pre-catalytic quaternary complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
8XFV
 
 | |
4JZO
 
 | |
4JZC
 
 | | Angiopoietin-2 fibrinogen domain TAG mutant | | Descriptor: | Angiopoietin-2 | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-04-02 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
9C82
 
 | | Structure of human ULK1C:PI3KC3-C1 supercomplex | | Descriptor: | Beclin 1-associated autophagy-related key regulator, Beclin-1, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
