2DWN
 
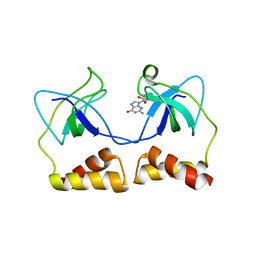 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
348D
 
 | |
6QLD
 
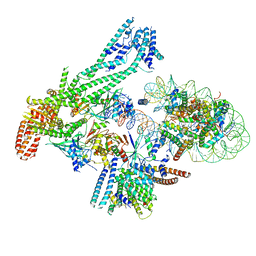 | | Structure of inner kinetochore CCAN-Cenp-A complex | | Descriptor: | DNA (125-MER), Histone H2A.1, Histone H2B.1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
5VL9
 
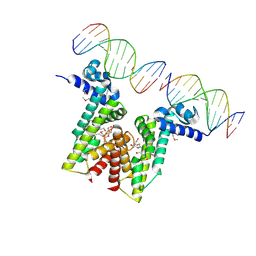 | | Crystal structure of EilR in complex with eilO DNA element | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*GP*TP*TP*GP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*TP*CP*CP*AP*AP*CP*TP*TP*TP*C)-3'), HEXANE-1,6-DIOL, ... | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
8D7Z
 
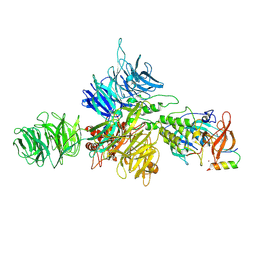 | | Cereblon-DDB1 bound to CC-92480 and Ikaros ZF1-2-3 | | Descriptor: | DNA damage-binding protein 1, DNA-binding protein Ikaros, Mezigdomide, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
1BN9
 
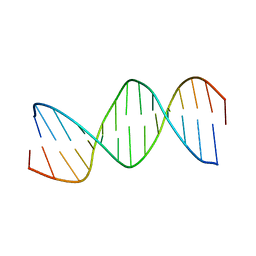 | | RESPONSE ELEMENT OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB BETA | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*CP*AP*TP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*TP*GP*TP*AP*GP*GP*TP*CP*AP*G)-3') | | Authors: | Castagne, C, Terenzi, H, Zakin, M.M, Delepierre, M. | | Deposit date: | 1998-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan nuclear receptor rev-erb beta response element by 1H, 31P NMR and molecular simulation
Biochimie, 82, 2000
|
|
3WVP
 
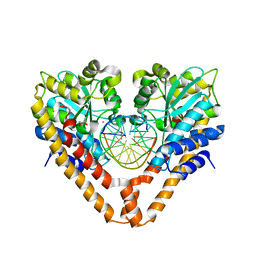 | | Time-Resolved Crystal Structure of HindIII with 60sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*A)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), DNA (5'-D(P*AP*GP*CP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-06-02 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6KIV
 
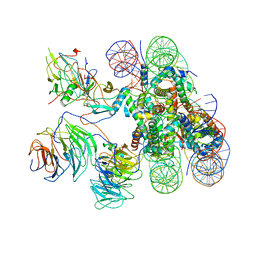 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIX
 
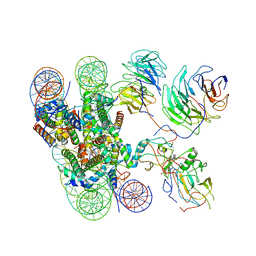 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
7LYA
 
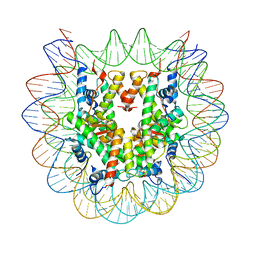 | | Cryo-EM structure of the human nucleosome core particle with linked histone proteins H2A and H2B | | Descriptor: | DNA (146-MER), DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
6KIU
 
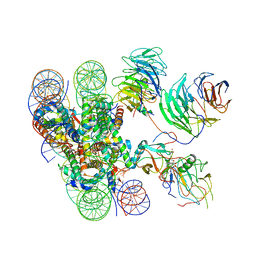 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIZ
 
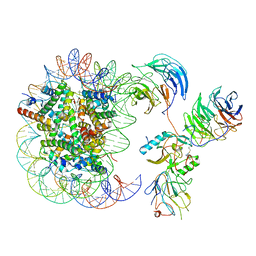 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
3WVK
 
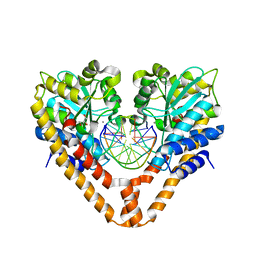 | | Time-Resolved Crystal Structure of HindIII with 230sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*A)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), DNA (5'-D(P*AP*GP*CP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-05-22 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
349D
 
 | |
1VTO
 
 | |
1I6V
 
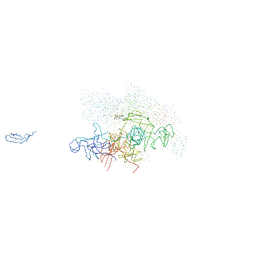 | | THERMUS AQUATICUS CORE RNA POLYMERASE-RIFAMPICIN COMPLEX | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, MAGNESIUM ION, RIFAMPICIN, ... | | Authors: | Campbell, E.A, Korzheva, N, Mustaev, A, Murakami, K, Goldfarb, A, Darst, S.A. | | Deposit date: | 2001-03-05 | | Release date: | 2001-04-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural mechanism for rifampicin inhibition of bacterial rna polymerase.
Cell(Cambridge,Mass.), 104, 2001
|
|
1B96
 
 | |
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
351D
 
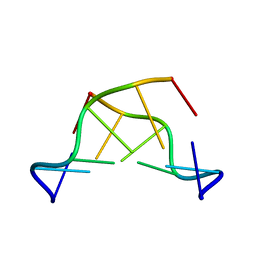 | |
1B95
 
 | |
7NL0
 
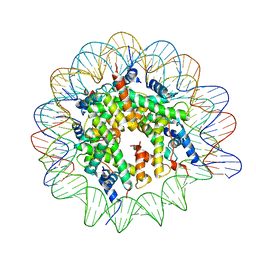 | | Cryo-EM structure of the Lin28B nucleosome core particle | | Descriptor: | DNA (131-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Roberts, G.A, Ozkan, B, Gachulincova, I, O Dwyer, M.R, Hall-Ponsele, E, Saxena, M, Robinson, P.J, Soufi, A. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dissecting OCT4 defines the role of nucleosome binding in pluripotency.
Nat.Cell Biol., 23, 2021
|
|
2C2J
 
 | | Crystal Structure Of The Dps92 From Deinococcus Radiodurans | | Descriptor: | DNA-BINDING STRESS RESPONSE PROTEIN, FE (III) ION, MAGNESIUM ION | | Authors: | Cuypers, M.G, Romao, C.V, Mitchell, E, McSweeney, S. | | Deposit date: | 2005-09-29 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the Dps2 from Deinococcus Radiodurans Reveals an Unusual Pore Profile with a Non-Specific Metal Binding Site.
J.Mol.Biol., 371, 2007
|
|
1DNX
 
 | | RNA/DNA DODECAMER R(G)D(CGTATACGC) WITH MAGNESIUM BINDING SITES | | Descriptor: | DNA/RNA (5'-R(*GP)-D(*CP*GP*TP*AP*TP*AP*CP*GP*C)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-12-16 | | Release date: | 2000-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes.
Nucleic Acids Res., 28, 2000
|
|
2C2F
 
 | | Dps from Deinococcus radiodurans | | Descriptor: | DNA-BINDING STRESS RESPONSE PROTEIN, FE (III) ION, GLYCEROL, ... | | Authors: | Romao, C.V, Mitchell, E, McSweeney, S. | | Deposit date: | 2005-09-28 | | Release date: | 2006-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Crystal Structure of Deinococcus Radiodurans Dps Protein (Dr2263) Reveals the Presence of a Novel Metal Centre in the N Terminus.
J.Biol.Inorg.Chem., 11, 2006
|
|
7R76
 
 | | Cryo-EM structure of DNMT5 in apo state | | Descriptor: | DNA repair protein Rad8, ZINC ION | | Authors: | Wang, J, Patel, D.J. | | Deposit date: | 2021-06-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
