6M96
 
 | |
8Q9R
 
 | | Crystal structure of MADS-box/MEF2D N-terminal domain bound to dsDNA and HDAC9 deacetylase binding motif | | Descriptor: | Histone deacetylase 9 (HDAC9) binding motif peptide: EVKQKLQEFLLSKS, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
5C7S
 
 | |
7MPT
 
 | | Brucella melitensis NrnC with bound Mg2+ | | Descriptor: | MAGNESIUM ION, NanoRNase C, PHOSPHATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
5CNO
 
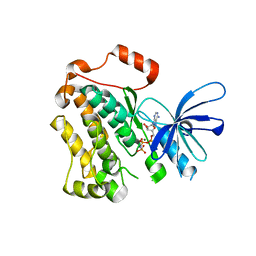 | | Crystal structure of the EGFR kinase domain mutant V924R | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kovacs, E, Das, R, Mirza, A, Jura, N, Barros, T, Kuriyan, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor.
Mol.Cell.Biol., 35, 2015
|
|
7XWZ
 
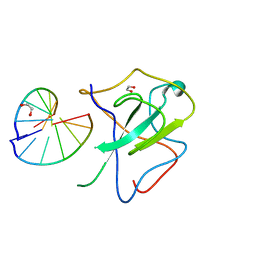 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
5FA7
 
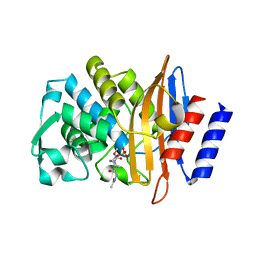 | | CTX-M-15 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-(acetamidocarbamoyl)-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
6MD5
 
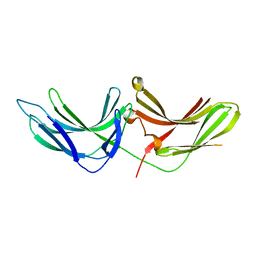 | |
5T8P
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to benzoxepin compound 2 | | Descriptor: | 6,7-dihydrothieno[4,5]oxepino[1,2-~{c}]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Smith, M.A, McEwan, P.A. | | Deposit date: | 2016-09-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5FQL
 
 | | Insights into Hunter syndrome from the structure of iduronate-2- sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demydchuk, M, Hill, C.H, Zhou, A, Bunkoczi, G, Stein, P.E, Marchesan, D, Deane, J.E, Read, R.J. | | Deposit date: | 2015-12-11 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into Hunter syndrome from the structure of iduronate-2-sulfatase.
Nat Commun, 8, 2017
|
|
8U32
 
 | | Crystal structure of PD-1 in complex with a Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2023-09-07 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure- and machine learning-guided engineering demonstrate that a non-canonical disulfide in an anti-PD-1 rabbit antibody does not impede antibody developability.
Mabs, 16, 2024
|
|
7MPU
 
 | | Brucella melitensis NrnC bound to pGG | | Descriptor: | 5'-phosphorylated GG, NanoRNase C, SULFATE ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural characterization of NrnC identifies unifying features of dinucleotidases.
Elife, 10, 2021
|
|
8QHK
 
 | | Crystal structure of reduced respiratory Complex I subunits NuoEF from Aquifex aeolicus bound to reduced 3-acetylpyridine adenine dinucleotide | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, ... | | Authors: | Wohlwend, D, Friedrich, T, Bucka, S. | | Deposit date: | 2023-09-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of 3-acetylpyridine adenine dinucleotide and ADP-ribose bound to the electron input module of respiratory complex I.
Structure, 32, 2024
|
|
5TDQ
 
 | | Crystal Structure of the GOLD domain of ACBD3 | | Descriptor: | Golgi resident protein GCP60 | | Authors: | McPhail, J.A, Burke, J.E. | | Deposit date: | 2016-09-19 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | The Molecular Basis of Aichi Virus 3A Protein Activation of Phosphatidylinositol 4 Kinase III beta , PI4KB, through ACBD3.
Structure, 25, 2017
|
|
7OKP
 
 | | Crystal structure of mouse CARM1 in complex with histone H3_13-22 K18 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.3, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
8TDF
 
 | |
7MPP
 
 | |
6MAJ
 
 | | HBO1 is required for the maintenance of leukaemia stem cells | | Descriptor: | 4-fluoro-N'-[(3-hydroxyphenyl)sulfonyl]-5-methyl[1,1'-biphenyl]-3-carbohydrazide, BRD1 protein, GLYCEROL, ... | | Authors: | Ren, B, Peat, T.S, Monahan, B, Dawson, M, Street, I. | | Deposit date: | 2018-08-27 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.139 Å) | | Cite: | HBO1 is required for the maintenance of leukaemia stem cells.
Nature, 577, 2020
|
|
5FYO
 
 | | Calcium-dependent phosphoinositol-specific phospholipase C from a Gram-negative bacterium, Pseudomonas sp, apo form, crystal form 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHOINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Norgaard, A, Segura, D.R, Blicher, T.H, Wilson, K.S. | | Deposit date: | 2016-03-08 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a calcium-dependent phosphoinositide-specific phospholipase C from Pseudomonas sp. 62186, the first from a Gram-negative bacterium.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6MAV
 
 | |
7OZQ
 
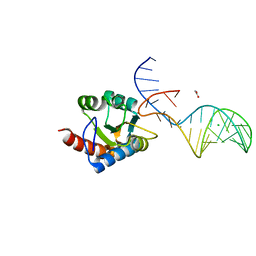 | | Crystal structure of archaeal L7Ae bound to eukaryotic kink-loop | | Descriptor: | 50S ribosomal protein L7Ae, ACETATE ION, CALCIUM ION, ... | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Eukaryotic Box C/D methylation machinery has two non-symmetric protein assembly sites.
Sci Rep, 11, 2021
|
|
7TTK
 
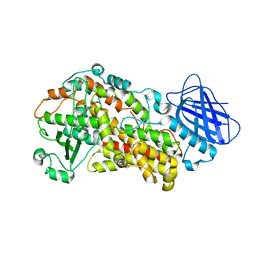 | | Stable-5-LOX | | Descriptor: | Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Gilbert, N.C, Newcomer, M.E. | | Deposit date: | 2022-02-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Stable-5-LOX "closed"
To Be Published
|
|
5G0C
 
 | | An unusual natural product primary sulfonamide: synthesis, carbonic anhydrase inhibition and protein x-ray structure of Psammaplin C | | Descriptor: | CARBONIC ANHYDRASE 2, ETHANOL, SODIUM ION, ... | | Authors: | Mujumdar, P, Supuran, C.T, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2016-03-17 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | An Unusual Natural Product Primary Sulfonamide: Synthesis, Carbonic Anhydrase Inhibition and Protein X-Ray Structures of Psammaplin C.
J.Med.Chem., 59, 2016
|
|
7P75
 
 | |
5T90
 
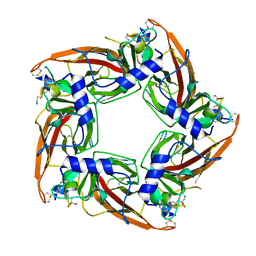 | | Structural mechanisms for alpha-conotoxin selectivity at the human alpha3beta4 nicotinic acetylcholine receptor | | Descriptor: | Acetylcholine-binding protein, LsIA | | Authors: | Abraham, N, Healy, M, Ragnarsson, L, Brust, A, Alewood, P, Lewis, R. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms for alpha-conotoxin activity at the human alpha 3 beta 4 nicotinic acetylcholine receptor.
Sci Rep, 7, 2017
|
|
