7LOH
 
 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail | | Descriptor: | Transmembrane protein gp41 | | Authors: | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
6GAK
 
 | |
5CVV
 
 | | coniferyl alcohol bound monolignol 4-O-methyltransferase 9 | | Descriptor: | (Iso)eugenol O-methyltransferase, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cai, Y, Liu, C.-J. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of coniferyl alcohol bound monolignol 4-O-methyltransferase 9 at 1.73 Angstroms resolution
To Be Published
|
|
3K83
 
 | | Crystal Structure Analysis of a Biphenyl/Oxazole/Carboxypyridine alpha-ketoheterocycle Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | 1-DODECANOL, 6-[2-(3-biphenyl-4-ylpropanoyl)-1,3-oxazol-5-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
4PZH
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
4CH9
 
 | | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK4 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, KELCH-LIKE PROTEIN 3, ... | | Authors: | Sorrell, F.J, Schumacher, F.R, Kurz, T, Alessi, D.R, Newman, J, Goubin, S, Chalk, R, Kopec, J, Tallant, C, Williams, E, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-11-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and Biochemical Characterisation of the Klhl3-Wnk Kinase Interaction Important in Blood Pressure Regulation.
Biochem.J., 460, 2014
|
|
6MH7
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SKT-68, a 1,4,5-trisubstituted imidazole analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2018-09-17 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular Basis for the N-Terminal Bromodomain-and-Extra-Terminal-Family Selectivity of a Dual Kinase-Bromodomain Inhibitor.
J.Med.Chem., 61, 2018
|
|
4LJH
 
 | | Crystal Structure of Pseudomonas aeruginosa Lectin LecA Complexed with 1-Methyl-3-indolyl-b-D-galactopyranoside at 1.45 A Resolution | | Descriptor: | 1-methyl-1H-indol-3-ol, CALCIUM ION, PA-I galactophilic lectin, ... | | Authors: | Kadam, R.U, Stocker, A, Reymond, J.L. | | Deposit date: | 2013-07-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CH-pi "T-Shape" Interaction with Histidine Explains Binding of Aromatic Galactosides to Pseudomonas aeruginosa Lectin LecA
Acs Chem.Biol., 8, 2013
|
|
5IQ7
 
 | | Crystal structure of 10E8-S74W Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8-S74W Heavy Chain, 10E8-S74W Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2869 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
4BJP
 
 | | Crystal structure of E. coli penicillin binding protein 3 | | Descriptor: | 1,2-ETHANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sauvage, E, Joris, M, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2013-04-19 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 3 (Pbp3) from Escherichia Coli.
Plos One, 9, 2014
|
|
5EQJ
 
 | | Crystal structure of the two-subunit tRNA m1A58 methyltransferase from Saccharomyces cerevisiae | | Descriptor: | tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRM61, tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit TRM6 | | Authors: | Zhu, Y, Wang, M, Wang, C, Fan, X, Jiang, X, Teng, M, Li, X. | | Deposit date: | 2015-11-13 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the two-subunit tRNA m(1)A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
7GCH
 
 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | Descriptor: | 1,1,1-TRIFLUORO-3-((N-ACETYL)-L-LEUCYLAMIDO)-4-PHENYL-BUTAN-2-ONE(N-ACETYL-L-LEUCYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | Authors: | Brady, K, Ringe, D, Abeles, R.H. | | Deposit date: | 1990-04-06 | | Release date: | 1990-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
7Z5W
 
 | | ROS1 with AstraZeneca ligand 1 | | Descriptor: | Proto-oncogene tyrosine-protein kinase ROS, SULFATE ION, ~{N}-[6-methyl-2-[(2~{S})-2-[3-(3-methylpyrazin-2-yl)-1,2-oxazol-5-yl]pyrrolidin-1-yl]pyrimidin-4-yl]-1,3-thiazol-2-amine | | Authors: | Hargreaves, D. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Virtual Screening in the Cloud Identifies Potent and Selective ROS1 Kinase Inhibitors.
J.Chem.Inf.Model., 62, 2022
|
|
6CFS
 
 | |
3KDB
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10006 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Lafont, V, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
4MWO
 
 | | Crystal structure of human mitochondrial 5'(3')-deoxyribonucleotidase in complex with the inhibitor CPB-T | | Descriptor: | 1-{3,5-O-[(4-carboxyphenyl)(phosphono)methylidene]-2-deoxy-beta-D-threo-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, 5'(3')-deoxyribonucleotidase, mitochondrial, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2013-09-25 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformationally constrained nucleoside phosphonic acids - potent inhibitors of human mitochondrial and cytosolic 5'(3')-nucleotidases.
Org.Biomol.Chem., 12, 2014
|
|
5Z5F
 
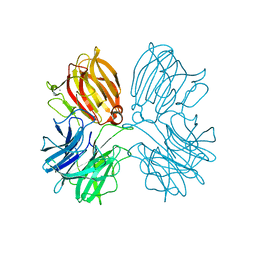 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose | | Descriptor: | Beta-xylosidase, CALCIUM ION, beta-L-arabinofuranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
1AWV
 
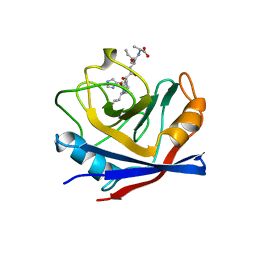 | | CYPA COMPLEXED WITH HVGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
1SZU
 
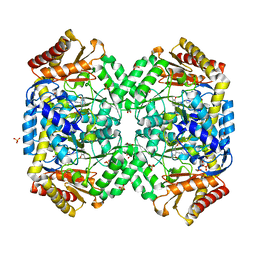 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
1GK9
 
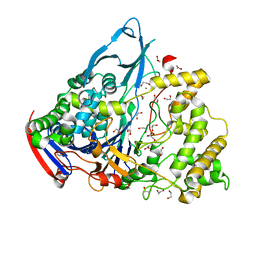 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme-Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
4GEM
 
 | | Crystal structure of Zucchini (K171A) | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, ZINC ION | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
5IJR
 
 | | X-ray structure of neuropilin-1 b1 domain complexed with Arg-1 ligand. | | Descriptor: | DIMETHYL SULFOXIDE, L-HOMOARGININE, Neuropilin-1 | | Authors: | Fotinou, C, Rana, R, Djordjevic, S, Yelland, T. | | Deposit date: | 2016-03-02 | | Release date: | 2017-03-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Architecture and hydration of the arginine-binding site of neuropilin-1.
FEBS J., 285, 2018
|
|
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
1BBO
 
 | |
4KP8
 
 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with 3-[(Pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(pyrimidin-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2013-05-13 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Benzenesulfonamides with pyrimidine moiety as inhibitors of human carbonic anhydrases I, II, VI, VII, XII, and XIII
Bioorg.Med.Chem., 21, 2013
|
|
