4X6S
 
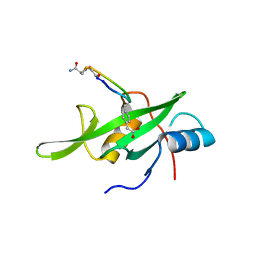 | | Grb7 SH2 domain with phosphotyrosine mimetic inhibitor peptide | | Descriptor: | Growth factor receptor-bound protein 7, Phosphotyrosine mimetic inhibitor peptide G7-TEM1 | | Authors: | Watson, G.M, Panjikar, S, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-09-23 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
J.Med.Chem., 58, 2015
|
|
4X1N
 
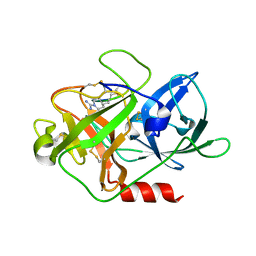 | | The crystal structure of mupain-1-16 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4X2K
 
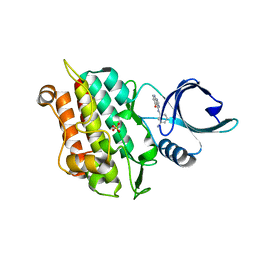 | | Selection of fragments for kinase inhibitor design: decoration is key | | Descriptor: | 4-[(3-aminophenyl)amino]pyrido[2,3-d]pyrimidin-5(6H)-one, SULFATE ION, TGF-beta receptor type-1 | | Authors: | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | Deposit date: | 2014-11-26 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
4WLX
 
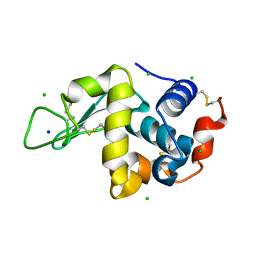 | | High pressure protein crystallography of hen egg white lysozyme at 280 MPa | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Yamada, H, Nagae, T, Watanabe, N. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-08 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-pressure protein crystallography of hen egg-white lysozyme
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WM3
 
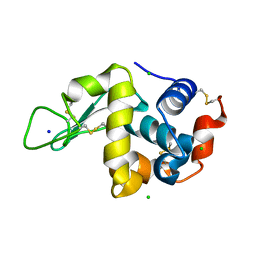 | | High pressure protein crystallography of hen egg white lysozyme at 710 MPa | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Yamada, H, Nagae, T, Watanabe, N. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-08 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-pressure protein crystallography of hen egg-white lysozyme
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4X6M
 
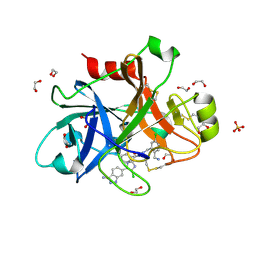 | |
4X6N
 
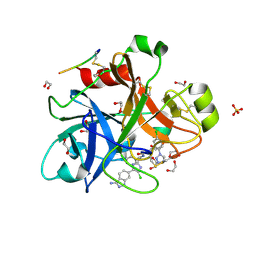 | |
4X7J
 
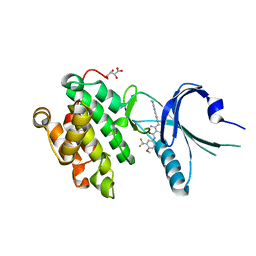 | | Co-crystal Structure of PERK with 2-amino-N-[4-methoxy-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-(methylamino)quinazolin-6-yl]benzamide inhibitor | | Descriptor: | 2-amino-N-[4-methoxy-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-(methylamino)quinazolin-6-yl]benzamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3, L(+)-TARTARIC ACID | | Authors: | Shaffer, P.L, Long, A.M, Chen, H. | | Deposit date: | 2014-12-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of 1H-Pyrazol-3(2H)-ones as Potent and Selective Inhibitors of Protein Kinase R-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 58, 2015
|
|
4WNV
 
 | | Human Cytochrome P450 2D6 Quinine Complex | | Descriptor: | Cytochrome P450 2D6, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4WPD
 
 | |
4WR9
 
 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WRC
 
 | | Crystal Structure of Surfactant Protein-A DEDN Mutant (E171D/P175E/R197N/K203D) | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
to be published
|
|
4WMZ
 
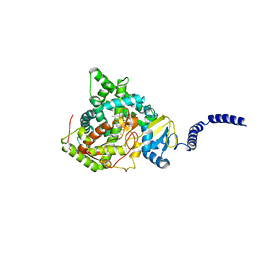 | | S. cerevisiae CYP51 complexed with fluconazole in the active site | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Wilbanks, S.M, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 59, 2015
|
|
4WTW
 
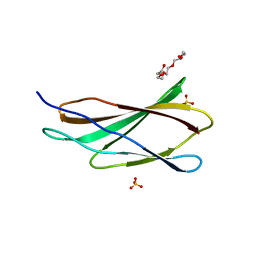 | | Crystal structure of the third FnIII domain of integrin beta4 | | Descriptor: | 1,2-ETHANEDIOL, Integrin beta-4, PENTAETHYLENE GLYCOL, ... | | Authors: | Alonso-Garcia, N, Urien, H, de Pereda, J.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin alpha6beta4
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4YUD
 
 | |
9EX1
 
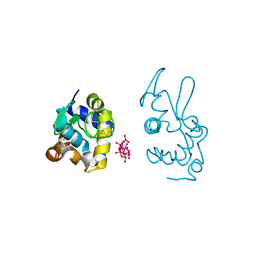 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure B) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9B8U
 
 | |
9CMZ
 
 | |
9EX0
 
 | | X-ray structure of a polyoxidovanadate/lysozyme adduct obtained when the protein is treated with [VIVO(acac)2] in 1.1 M NaCl, 0.1 M sodium acetate at pH 4.0 (Structure A) | | Descriptor: | CHLORIDE ION, Lysozyme C, Polyoxidovanadate complex, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2024-04-05 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-Covalent and Covalent Binding of New Mixed-Valence Cage-like Polyoxidovanadate Clusters to Lysozyme.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9LPR
 
 | |
8ZMN
 
 | | Crystal structure of ANTXR1 | | Descriptor: | Anthrax toxin receptor 1, SODIUM ION | | Authors: | Zheng, H, Hu, J, Fu, L, Chen, R. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray structure of Anthrax toxin receptor 1 APO from Rattus norvegicus
To Be Published
|
|
9CTX
 
 | | X-ray crystal structure of multi-drug resistant HIV-1 protease (P51) in complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, GLYCEROL, Protease | | Authors: | Hayashi, H, Yedidi, R, Bulut, H, Das, D, Mitsuya, H. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Secondary amino-acid substitutions contribute to the emergence of HIV protease inhibitor resistance as directly as primary amino-acid substitutions.
To Be Published
|
|
9EZ3
 
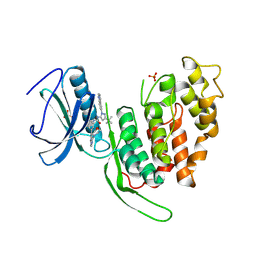 | | Crystal structure of human CLK3 bound to RN129 | | Descriptor: | 1-(5-~{tert}-butyl-2-quinolin-6-yl-pyrazol-3-yl)-3-[3-(4-morpholin-4-yl-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl)phenyl]urea, Dual specificity protein kinase CLK3, SULFATE ION | | Authors: | Kraemer, A, Raig, N, Knapp, S. | | Deposit date: | 2024-04-10 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human CLK3 bound to RN129
To Be Published
|
|
9FJO
 
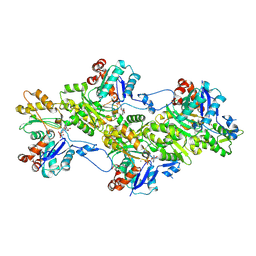 | | Structure of the undecorated pointed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Boiero Sanders, M, Oosterheert, W, Hofnagel, O, Bieling, P, Raunser, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Phalloidin and DNase I-bound F-actin pointed end structures reveal principles of filament stabilization and disassembly.
Nat Commun, 15, 2024
|
|
9INS
 
 | |
