1GD7
 
 | | CRYSTAL STRUCTURE OF A BIFUNCTIONAL PROTEIN (CSAA) WITH EXPORT-RELATED CHAPERONE AND TRNA-BINDING ACTIVITIES. | | Descriptor: | CSAA PROTEIN | | Authors: | Shibata, T, Inoue, Y, Vassylyev, D.G, Kawaguchi, S, Yokoyama, S, Muller, J, Linde, D, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the ttCsaA protein: an export-related chaperone from Thermus thermophilus.
EMBO J., 20, 2001
|
|
3JVF
 
 | | Crystal structure of an Interleukin-17 receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ely, L.K, Garcia, K.C. | | Deposit date: | 2009-09-16 | | Release date: | 2009-10-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of receptor sharing by interleukin 17 cytokines.
Nat.Immunol., 10, 2009
|
|
6NVO
 
 | | Crystal structure of Pseudomonas putida nuclease MPE | | Descriptor: | MANGANESE (II) ION, Nuclease MPE | | Authors: | Goldgur, Y, Shuman, S, Ejaz, A. | | Deposit date: | 2019-02-05 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Activity and structure ofPseudomonas putidaMPE, a manganese-dependent single-strand DNA endonuclease encoded in a nucleic acid repair gene cluster.
J.Biol.Chem., 294, 2019
|
|
6BWL
 
 | | X-ray structure of Pal from Bacillus thuringiensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Delvaux, N.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular architectures of Pen and Pal: Key enzymes required for CMP-pseudaminic acid biosynthesis in Bacillus thuringiensis.
Protein Sci., 27, 2018
|
|
5WRO
 
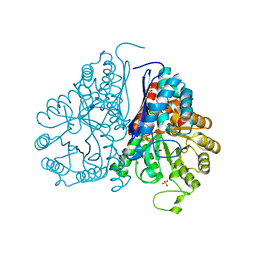 | | Crystal structure of Drosophila enolase | | Descriptor: | CADMIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Zhang, Z, Shi, Z. | | Deposit date: | 2016-12-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Crystal structure of enolase from Drosophila melanogaster.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4LTR
 
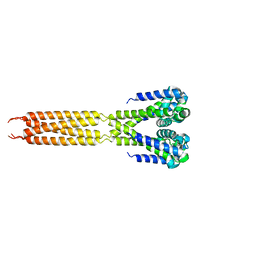 | | Bacterial sodium channel, His245Gly mutant, I222 space group | | Descriptor: | Ion transport protein | | Authors: | Shaya, D, Findeisen, F, Abderemane-Ali, F, Arrigoni, C, Wong, S, Reddy Nurva, S, Loussouarn, G, Minor, D.L. | | Deposit date: | 2013-07-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.8 Å) | | Cite: | Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels.
J.Mol.Biol., 426, 2014
|
|
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2CJ4
 
 | |
5WSX
 
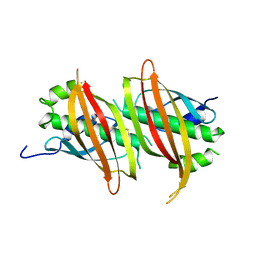 | | The crystal structure of SAV606 | | Descriptor: | Uncharacterized protein | | Authors: | Chisuga, T, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of the dual-function thioesterase SAV606 unravels the mechanism of Michael addition of glycine to an alpha , beta-unsaturated thioester.
J. Biol. Chem., 292, 2017
|
|
3G4Y
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloromethyl benzene bound to the XE4 cavity | | Descriptor: | (chloromethyl)benzene, CARBON MONOXIDE, GLOBIN-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
1GAE
 
 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
2HCB
 
 | | Structure of AMPPCP-bound DnaA from Aquifex aeolicus | | Descriptor: | Chromosomal replication initiator protein dnaA, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Erzberger, J.P, Mott, M.L, Berger, J.M. | | Deposit date: | 2006-06-15 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3G53
 
 | | Ligand migration and cavities within scapharca dimeric hemoglobin: wild type with co bound to heme and chloropropyl benzene bound to the XE4 cavity | | Descriptor: | (3-chloropropyl)benzene, CARBON MONOXIDE, Globin-1, ... | | Authors: | Knapp, J.E, Pahl, R, Cohen, J, Nichols, J.C, Schulten, K, Gibson, Q.H, Srajer, V, Royer Jr, W.E. | | Deposit date: | 2009-02-04 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand migration and cavities within Scapharca Dimeric HbI: studies by time-resolved crystallo-graphy, Xe binding, and computational analysis.
Structure, 17, 2009
|
|
1GGO
 
 | | T453A MUTANT OF PYRUVATE, PHOSPHATE DIKINASE | | Descriptor: | PROTEIN (PYRUVATE, PHOSPHATE DIKINASE), SULFATE ION | | Authors: | Li, Z, Herzberg, O. | | Deposit date: | 2000-08-29 | | Release date: | 2001-01-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of domain-domain docking sites within Clostridium symbiosum pyruvate phosphate dikinase by amino acid replacement.
J.Biol.Chem., 275, 2000
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | Descriptor: | FAB Heavy chain, FAB Light chain | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2H3K
 
 | | Solution Structure of the first NEAT domain of IsdH | | Descriptor: | Haptoglobin-binding surface anchored protein | | Authors: | Pilpa, R.M, Fadeev, E.A, Villareal, V.A, Wong, M.A, Phillips, M, Clubb, R.T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus.
J.Mol.Biol., 360, 2006
|
|
5CSS
 
 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilum with glycerol 3-phosphate | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Kim, K.R, Park, J.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
3DZD
 
 | | Crystal structure of sigma54 activator NTRC4 in the inactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SODIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-07-29 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
2DYP
 
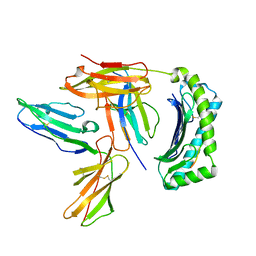 | | Crystal Structure of LILRB2(LIR2/ILT4/CD85d) complexed with HLA-G | | Descriptor: | 9 Mer Peptide From Histone H2A.x, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shiroishi, M, Kuroki, K, Rasubala, L, Kohda, D, Maenaka, K. | | Deposit date: | 2006-09-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d)
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1GC9
 
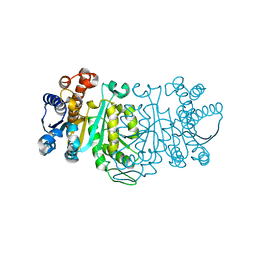 | | THE CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS 3-ISOPROPYLMALATE DEHYDROGENASE MUTATED AT 172TH FROM ALA TO GLY | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE | | Authors: | Qu, C, Akanuma, S, Tanaka, N, Moriyama, H, Oshima, T. | | Deposit date: | 2000-07-28 | | Release date: | 2000-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, X-ray crystallography, molecular modelling and thermal stability studies of mutant enzymes at site 172 of 3-isopropylmalate dehydrogenase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3QW9
 
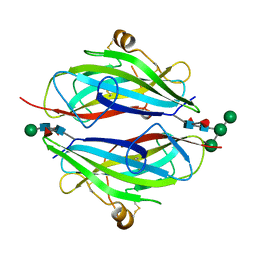 | | Crystal structure of betaglycan ZP-C domain | | Descriptor: | Transforming growth factor beta receptor type 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lin, S.J, Jardetzky, T.S. | | Deposit date: | 2011-02-27 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of betaglycan zona pellucida (ZP)-C domain provides insights into ZP-mediated protein polymerization and TGF-{beta} binding.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3KNG
 
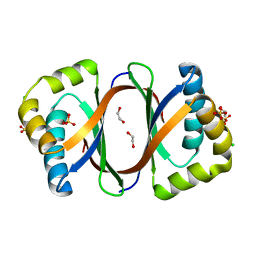 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.9 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-11-12 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3QWQ
 
 | |
1FX0
 
 | |
2IFW
 
 | | Crystal structure of scytalido-glutamic peptidase with a transition state analog inhibitor | | Descriptor: | ACETIC ACID, GLYCEROL, Heptapeptide, ... | | Authors: | Pillai, B, Cherney, M.M, Hiraga, K, Takada, K, Oda, K, James, M.N. | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of scytalidoglutamic peptidase with its first potent inhibitor provides insights into substrate specificity and catalysis.
J.Mol.Biol., 365, 2007
|
|
