8FSW
 
 | |
8FSU
 
 | |
5JB9
 
 | | Crystal structure of factor IXa K98T variant in complex with PPACK | | Descriptor: | CALCIUM ION, Coagulation factor IX, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Kristensen, L.H, Brandstetter, H. | | Deposit date: | 2016-04-13 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Releasing the brakes in coagulation Factor IXa by co-operative maturation of the substrate-binding site.
Biochem.J., 473, 2016
|
|
6WC2
 
 | | Crystal Structure of a Ternary MEF2 Chimera/NKX2-5/myocardin enhancer DNA Complex | | Descriptor: | Homeobox protein Nkx-2.5, MEF2 Chimera,Myocyte-specific enhancer factor 2B,Myocyte-specific enhancer factor 2A, Myocardin Enhancer DNA | | Authors: | Lei, X, Chen, L. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Ternary Complexes of MEF2 and NKX2-5 Bound to DNA Reveal a Disease Related Protein-Protein Interaction Interface.
J.Mol.Biol., 432, 2020
|
|
8FSV
 
 | |
8FST
 
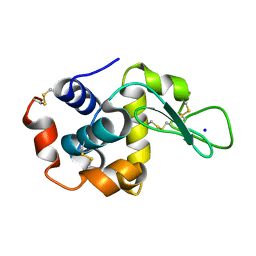 | |
8HY7
 
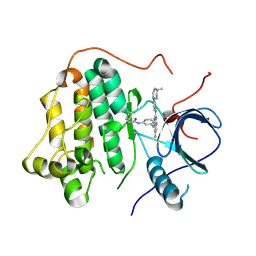 | | EGFR kinase domain mutant "TMLR" with compound 28f | | Descriptor: | Epidermal growth factor receptor, ~{N}-[3-[[6-fluoranyl-2-[[4-(4-methylpiperazin-1-yl)phenyl]amino]quinazolin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Wang, Y.X, Wu, C.Y. | | Deposit date: | 2023-01-06 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The crystal structure of EGFR kinase domain(T790M/L858R) in complex with compound 28f
To Be Published
|
|
4ZFL
 
 | |
5JAW
 
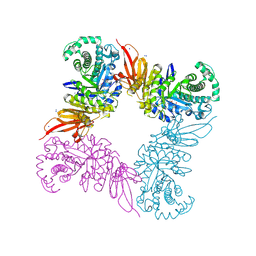 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1S,2S,3S,4S,5R,6R)-5-amino-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2016-04-12 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Towards broad spectrum activity-based glycosidase probes: synthesis and evaluation of deoxygenated cyclophellitol aziridines.
Chem. Commun. (Camb.), 53, 2017
|
|
8FT0
 
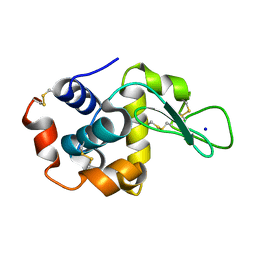 | |
8FSY
 
 | |
8FSX
 
 | |
8FT3
 
 | |
5JC9
 
 | | Structure of the Escherichia coli ribosome with the U1052G mutation in the 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-14 | | Release date: | 2016-07-06 | | Last modified: | 2016-08-03 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZH7
 
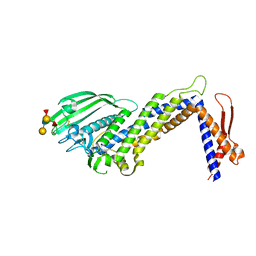 | | Structural basis of Lewisb antigen binding by the Helicobacter pylori adhesin BabA | | Descriptor: | Outer membrane protein-adhesin, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose | | Authors: | Howard, T, Hage, N, Phillips, C, Brassington, C.A, Debreczeni, J, Overman, R, Gellert, P, Stolnik, S, Winkler, G.S, Falcone, F.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA.
Sci Adv, 1, 2015
|
|
8I34
 
 | | The crystal structure of EPD-BCP1 from a marine sponge | | Descriptor: | (2~{Z},4~{E},6~{E},8~{E},10~{E},12~{E},14~{E},16~{E})-4,8,13,17-tetramethyl-3-oxidanyl-19-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]-1-[(1~{R},4~{S})-1,2,2-trimethyl-4-oxidanyl-cyclopentyl]nonadeca-2,4,6,8,10,12,14,16-octaen-18-yn-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASTAXANTHIN, ... | | Authors: | Shomura, Y, Kawasaki, S. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | An ependymin-related blue carotenoprotein decorates marine blue sponge.
J.Biol.Chem., 299, 2023
|
|
8IC1
 
 | | endo-alpha-D-arabinanase EndoMA1 D51N mutant from Microbacterium arabinogalactanolyticum in complex with arabinooligosaccharides | | Descriptor: | (3~{a}~{S},5~{R},6~{R},6~{a}~{S})-5-(hydroxymethyl)-2,2-dimethyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]dioxol-6-ol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, J, Nakashima, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8FYZ
 
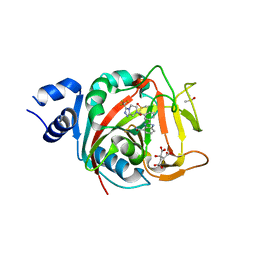 | | Crystal structure of human PARP1 ART domain bound to inhibitor UKTT10 (compound 13) | | Descriptor: | (2P)-2-{3-[(4R)-3-(trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazine-7(8H)-carbonyl]phenyl}-1H-benzimidazole-4-carboxamide, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Novel modifications of PARP inhibitor veliparib increase PARP1 binding to DNA breaks.
Biochem.J., 481, 2024
|
|
5JGZ
 
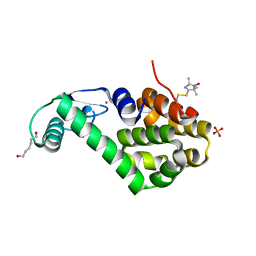 | | Spin-Labeled T4 Lysozyme Construct T151V1 | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
8FT1
 
 | |
6WGB
 
 | | Crystal structure of the fab portion of dupilumab | | Descriptor: | Dupilumab Fab heavy chain, Dupilumab Fab light chain | | Authors: | Druzina, Z, Atwell, S, Pustilnik, A, Antonysamy, S, Ho, C, Lieu, R, Hendle, J, Benach, J, Wang, J. | | Deposit date: | 2020-04-05 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
8IIA
 
 | |
8FZ8
 
 | | Structure of cytochrome P450sky2 | | Descriptor: | Cytochrome P450, OCTANOIC ACID (CAPRYLIC ACID), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murarka, V.C, Poulos, T.L. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Biosynthesis of a new skyllamycin in Streptomyces nodosus : a cytochrome P450 forms an epoxide in the cinnamoyl chain.
Org.Biomol.Chem., 22, 2024
|
|
8IC7
 
 | | exo-beta-D-arabinofuranosidase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with beta-D-arabinofuranose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8FK9
 
 | | Crystal Structure of the Tick Evasin EVA-ACA1001 Complexed to Human Chemokine CCL16 | | Descriptor: | C-C motif chemokine 16, Evasin P991 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of chemokine recognition by the class A3 tick evasin EVA-ACA1001.
Protein Sci., 33, 2024
|
|
