8EL2
 
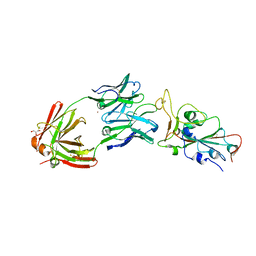 | | SARS-CoV-2 RBD bound to neutralizing antibody Fab ICO-hu23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ICO-hu23 Heavy Chain, Fab ICO-hu23 Light Chain, ... | | Authors: | Besaw, J.E, Kuo, A, Morizumi, T, Ernst, O.P. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
8ELJ
 
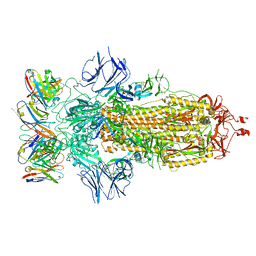 | | SARS-CoV-2 spike glycoprotein in complex with the ICO-hu23 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ICO-hu23 antibody Fab heavy chain, ... | | Authors: | Yee, A.W, Morizumi, T, Kim, K, Kuo, A, Ernst, O.P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing humanized SARS-CoV-2 antibody binds to a conserved epitope on Spike and provides antiviral protection through inhalation-based delivery in non-human primates.
Plos Pathog., 19, 2023
|
|
8EM5
 
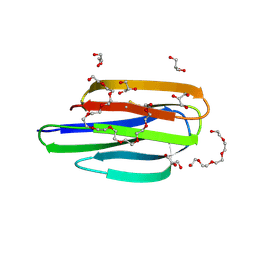 | | Mycobacterium thermoresistible MmpS5 | | Descriptor: | DODECAETHYLENE GLYCOL, GLYCEROL, IODIDE ION, ... | | Authors: | Cuthbert, B.J, Goulding, C.W. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Mycobacterium thermoresistibile MmpS5 reveals a conserved disulfide bond across mycobacteria.
Metallomics, 16, 2024
|
|
8F0I
 
 | |
8EZV
 
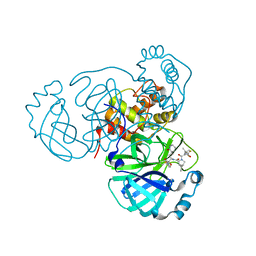 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8EZZ
 
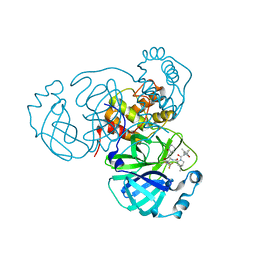 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a2 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-difluoroazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8F2D
 
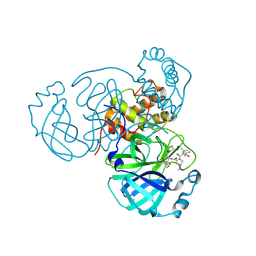 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML4006a | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopiperidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8EQF
 
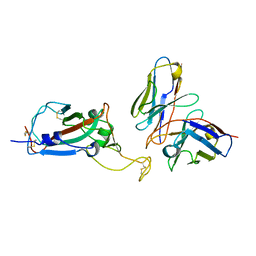 | | cryoEM structure of a broadly neutralizing anti-SARS-CoV-2 antibody STI-9167 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Bajic, G. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | An in vitro experimental pipeline to characterize the epitope of a SARS-CoV-2 neutralizing antibody.
Mbio, 15, 2024
|
|
8F02
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML2006a4 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(3,3-dimethylazetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-01 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8F2C
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML3006a | | Descriptor: | (1R,2S,5S)-N-[(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-(2-oxopyrrolidin-1-yl)butan-2-yl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-11-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
8EKD
 
 | |
8FD5
 
 | | Nucleocapsid monomer structure from SARS-CoV-2 | | Descriptor: | Nucleoprotein | | Authors: | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | Deposit date: | 2022-12-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (4.57 Å) | | Cite: | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
8FWX
 
 | |
8FY7
 
 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase nsp5, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-1-[(3S)-2-oxopyrrolidin-3-yl]but-3-en-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8FY6
 
 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | (1R,2S,5S)-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-N-{(2R)-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXC
 
 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
8GOP
 
 | | SARS-CoV-2 specific private TCR RLQ7 | | Descriptor: | SARS-CoV-2 specific private TCR RLQ7 alpha, SARS-CoV-2 specific private TCR RLQ7 beta | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
J.Biol.Chem., 299, 2023
|
|
8GOM
 
 | | SARS-CoV-2 specific private TCR RLQ7 in complex with RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, SARS-CoV-2 specific private TCR RLQ7 alpha, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Structural insights into protection against a SARS-CoV-2 spike variant by T cell receptor (TCR) diversity.
J.Biol.Chem., 299, 2023
|
|
8GON
 
 | |
6WXD
 
 | | SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
6WPS
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | Authors: | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-27 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
6WZQ
 
 | |
