8IBI
 
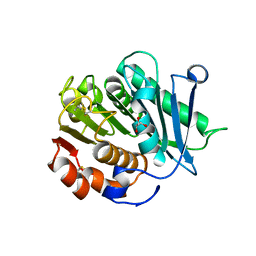 | | Inactive mutant of CtPL-H210S/F214I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
7S0H
 
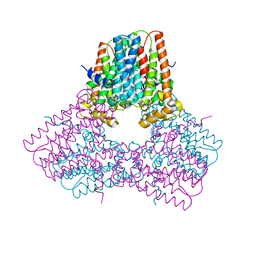 | |
7NDL
 
 | | Crystal structure of human GFAT-1 S205D | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Isoform 2 of Glutamine-fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Baumann, U, Denzel, M.S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
7S0A
 
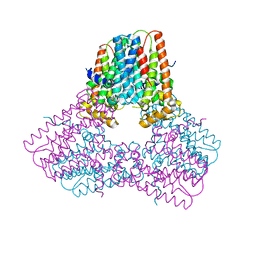 | |
3LQR
 
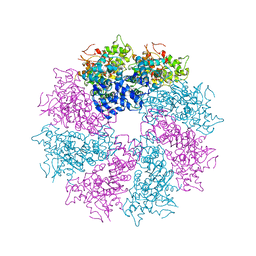 | | Structure of CED-4:CED-3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 4, MAGNESIUM ION | | Authors: | Qi, S, Pang, Y, Shi, Y, Yan, N. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.896 Å) | | Cite: | Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4.
Cell(Cambridge,Mass.), 141, 2010
|
|
4PG4
 
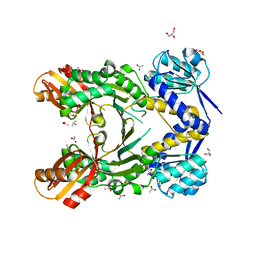 | |
3CFC
 
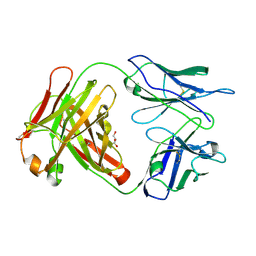 | | High-resolution structure of blue fluorescent antibody EP2-19G2 | | Descriptor: | BLUE FLUORESCENT ANTIBODY EP2-19G2-IGG2B HEAVY CHAIN, BLUE FLUORESCENT ANTIBODY EP2-19G2-KAPPA LIGHT CHAIN, GLYCEROL | | Authors: | Debler, E.W, Wilson, I.A. | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Deeply inverted electron-hole recombination in a luminescent antibody-stilbene complex.
Science, 319, 2008
|
|
4PUR
 
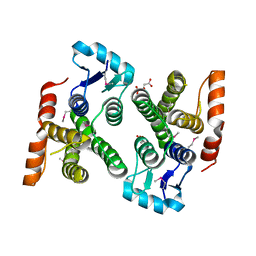 | | Crystal structure of MglA from Francisella tularensis | | Descriptor: | D-MALATE, Macrophage growth locus, subunit A | | Authors: | Cuthbert, B.J, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 2014-03-13 | | Release date: | 2015-06-24 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and Biochemical Characterization of the Francisella tularensis Pathogenicity Regulator, Macrophage Locus Protein A (MglA).
Plos One, 10, 2015
|
|
7S0M
 
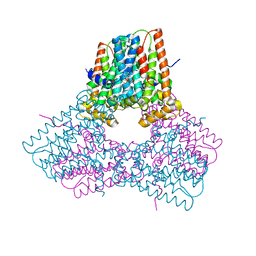 | |
8I5I
 
 | |
7S09
 
 | |
4PH4
 
 | | The crystal structure of Human VPS34 in complex with PIK-III | | Descriptor: | 4'-(cyclopropylmethyl)-N~2~-(pyridin-4-yl)-4,5'-bipyrimidine-2,2'-diamine, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2014-05-04 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo.
Nat.Cell Biol., 16, 2014
|
|
8CBX
 
 | | Crystal Structure of Anti-Cortisol Fab fragment | | Descriptor: | TETRAETHYLENE GLYCOL, anti-cortisol (17) Fab (heavy chain), anti-cortisol (17) Fab (light chain) | | Authors: | Eronen, V, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight to elucidate the binding specificity of the anti-cortisol Fab fragment with glucocorticoids.
J.Struct.Biol., 215, 2023
|
|
8I5H
 
 | |
7NIO
 
 | | Crystal structure of the SARS-CoV-2 helicase APO form | | Descriptor: | SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
2JU0
 
 | | Structure of Yeast Frequenin bound to PdtIns 4-kinase | | Descriptor: | CALCIUM ION, Calcium-binding protein NCS-1, Phosphatidylinositol 4-kinase PIK1 | | Authors: | Ames, J. | | Deposit date: | 2007-08-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1).
J.Biol.Chem., 282, 2007
|
|
8IBJ
 
 | | Inactive mutant of CtPL-H210S/F214I/N181A/F235L | | Descriptor: | PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
3LT8
 
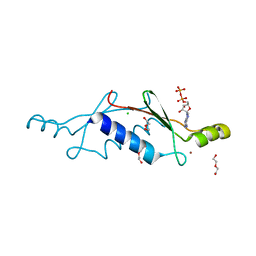 | | A non-biological ATP binding protein with a single point mutation (D65V), that contributes to optimized folding and ligand binding, crystallized in the presence of 100 mM ATP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-D65V, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
8CH6
 
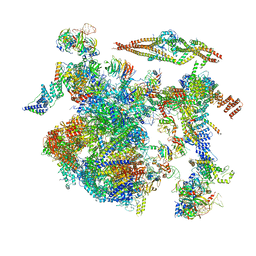 | | Structure of a late-stage activated spliceosome (BAqr) arrested with a dominant-negative Aquarius mutant (state B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Cretu, C, Schmitzova, J, Pena, V. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of catalytic activation in human splicing.
Nature, 617, 2023
|
|
8I8Y
 
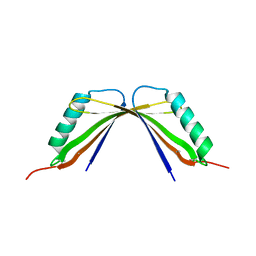 | | A mutant of the C-terminal complex of proteins 4.1G and NuMA | | Descriptor: | Engineered protein | | Authors: | Hu, X. | | Deposit date: | 2023-02-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined prediction and design reveals the target recognition mechanism of an intrinsically disordered protein interaction domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SGC
 
 | |
3D5R
 
 | | Crystal Structure of Efb-C (N138A) / C3d Complex | | Descriptor: | Complement C3, Fibrinogen-binding protein | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2008-05-16 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d.
Protein Sci., 17, 2008
|
|
8IAN
 
 | | Crystal structure of CtPL-H210S/F214I mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8CBZ
 
 | |
7NN0
 
 | | Crystal structure of the SARS-CoV-2 helicase in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SARS-CoV-2 helicase NSP13, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
