8DEW
 
 | |
6MRL
 
 | | Cucumber Leaf Spot Virus | | Descriptor: | CALCIUM ION, p41 | | Authors: | Sherman, M.B, Smith, T.J. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes.
J.Virol., 94, 2020
|
|
8EHG
 
 | | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
6NNB
 
 | |
6VPO
 
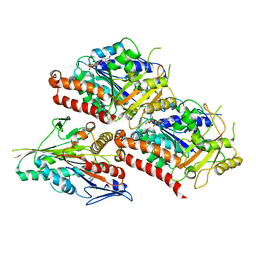 | | Cryo-EM structure of microtubule-bound KLP61F motor domain in the AMPPNP state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein Klp61F, ... | | Authors: | Bodrug, T, Wilson-Kubalek, E.M, Nithianantham, S, Debs, G, Sindelar, C.V, Milligan, R, Al-Bassam, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding.
Elife, 9, 2020
|
|
6U5O
 
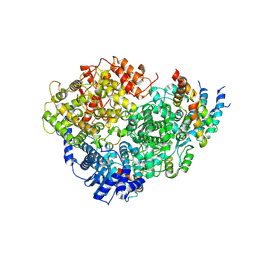 | | Structure of the Human Metapneumovirus Polymerase bound to the phosphoprotein tetramer | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L | | Authors: | Pan, J, Qian, X, Lattmann, S, Sahili, A.E, Yeo, T.H, Kalocsay, M, Fearns, R, Lescar, J. | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the human metapneumovirus polymerase phosphoprotein complex.
Nature, 577, 2020
|
|
4JIB
 
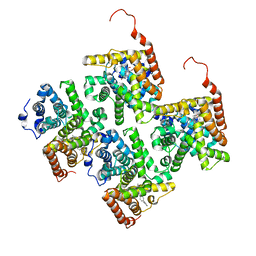 | | Crystal structure of of PDE2-inhibitor complex | | Descriptor: | 1-(2-hydroxyethyl)-3-(2-methylbutan-2-yl)-5-[4-(2-methyl-1H-imidazol-1-yl)phenyl]-6,7-dihydropyrazolo[4,3-e][1,4]diazepin-8(1H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Pandit, J. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of potent, selective, bioavailable phosphodiesterase 2 (PDE2) inhibitors active in an osteoarthritis pain model, Part I: Transformation of selective pyrazolodiazepinone phosphodiesterase 4 (PDE4) inhibitors into selective PDE2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8WZH
 
 | |
6XRE
 
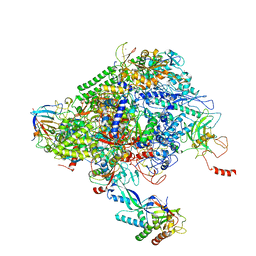 | | Structure of the p53/RNA polymerase II assembly | | Descriptor: | Cellular tumor antigen p53, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Liou, S.-H, Singh, S, Singer, R.H, Coleman, R.A, Liu, W. | | Deposit date: | 2020-07-12 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the p53/RNA polymerase II assembly.
Commun Biol, 4, 2021
|
|
6R7F
 
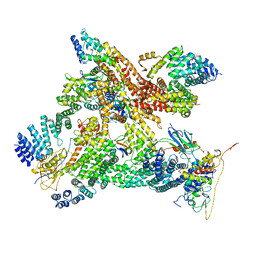 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7H
 
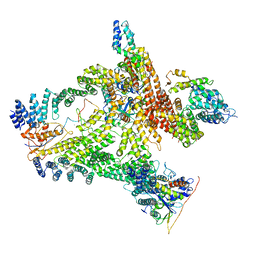 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7N
 
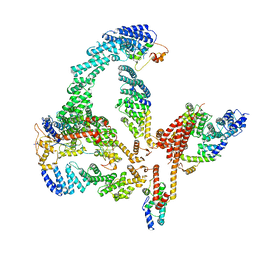 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7I
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R6H
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Morris, E.P, Faull, S.V, Lau, A.M.C, Politis, A, Beuron, F, Cronin, N. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6OMM
 
 | | Cryo-EM structure of formyl peptide receptor 2/lipoxin A4 receptor in complex with Gi | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Liu, H, de Waal, P.W, Zhou, X.E, Wang, L, Meng, X, Zhao, G, Kang, Y, Melcher, K, Xu, H.E, Zhang, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure of formylpeptide receptor 2-Gicomplex reveals insights into ligand recognition and signaling.
Nat Commun, 11, 2020
|
|
7SVR
 
 | |
6V21
 
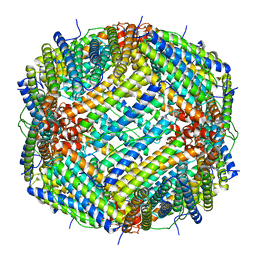 | |
7TDZ
 
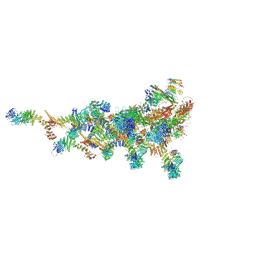 | |
6VAU
 
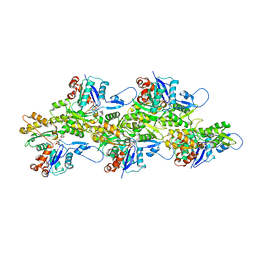 | | Bare actin filament from a partially cofilin-decorated sample | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2UZK
 
 | | Crystal structure of the human FOXO3a-DBD bound to DNA | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*AP*G)-3', FORKHEAD BOX PROTEIN O3A | | Authors: | Tsai, K.-L, Sun, Y.-J, Huang, C.-Y, Yang, J.-Y, Hung, M.-C, Hsiao, C.-D. | | Deposit date: | 2007-04-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Human Foxo3A-Dbd/DNA Complex Suggests the Effects of Post-Translational Modification.
Nucleic Acids Res., 35, 2007
|
|
6Z6W
 
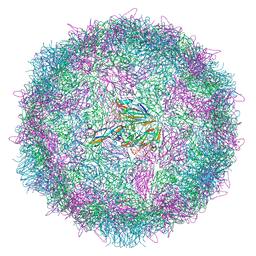 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) from a mammalian expression system. | | Descriptor: | Capsid proteins, VP0, VP1, ... | | Authors: | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mammalian expression of virus-like particles as a proof of principle for next generation polio vaccines.
NPJ Vaccines, 6, 2021
|
|
6VM1
 
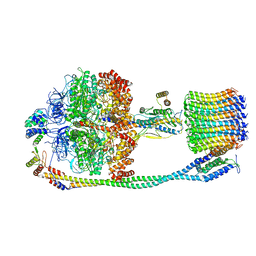 | | Chloroplast ATP synthase (C3, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6ZIT
 
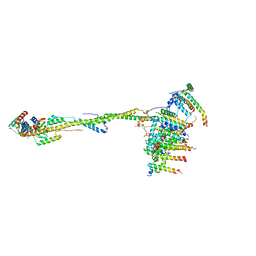 | | bovine ATP synthase Stator domain, state 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C2, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZIU
 
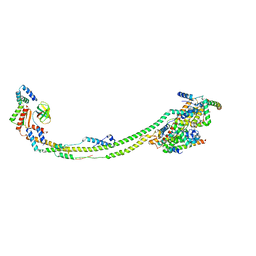 | | bovine ATP synthase stator domain, state 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase protein 8, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZIK
 
 | | bovine ATP synthase rotor domain, state 3 | | Descriptor: | ATP synthase F(0) complex subunit C2, mitochondrial, ATP synthase subunit delta, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
