3AQN
 
 | |
3AQL
 
 | |
3AVT
 
 | | Structure of viral RNA polymerase complex 1 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVW
 
 | | Structure of viral RNA polymerase complex 4 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVU
 
 | | Structure of viral RNA polymerase complex 2 | | Descriptor: | CALCIUM ION, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AVX
 
 | | Structure of viral RNA polymerase complex 5 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
6KR2
 
 | | Crystal structure of Dengue virus nonstructural protein NS5 (form 1) | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Wu, J, Lu, G, Ye, H.Q, Gong, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A conformation-based intra-molecular initiation factor identified in the flavivirus RNA-dependent RNA polymerase.
Plos Pathog., 16, 2020
|
|
6KR3
 
 | | Crystal structure of Dengue virus nonstructural protein NS5 (form 2) | | Descriptor: | GLYCEROL, Genome polyprotein, IODIDE ION, ... | | Authors: | Wu, J, Lu, G, Ye, H.Q, Gong, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | A conformation-based intra-molecular initiation factor identified in the flavivirus RNA-dependent RNA polymerase.
Plos Pathog., 16, 2020
|
|
9BDC
 
 | | Cryo-EM Structure of the TEFM bound Human Mitochondrial Transcription Elongation Complex in a Closed Fingers Domain Conformation | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Non-Template Strand DNA (NT27mt_+1T), ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2024-04-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
9BDD
 
 | | Cryo-EM Structure of Non-Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2024-04-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
9CGI
 
 | |
1KKS
 
 | | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression | | Descriptor: | 5'-R(*GP*GP*AP*AP*GP*GP*CP*CP*CP*UP*UP*UP*UP*CP*AP*GP*GP*GP*CP*CP*AP*CP*CP*C)-3' | | Authors: | Zanier, K, Luyten, I, Crombie, C, Muller, B, Schuemperli, D, Linge, J.P, Nilges, M, Sattler, M. | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression.
RNA, 8, 2002
|
|
7OEA
 
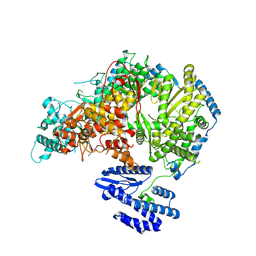 | | Lassa virus L protein bound to 3' promoter RNA (well-resolved polymerase core) [3END-CORE] | | Descriptor: | 3' vRNA, MAGNESIUM ION, RNA-directed RNA polymerase L, ... | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-02 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7OCH
 
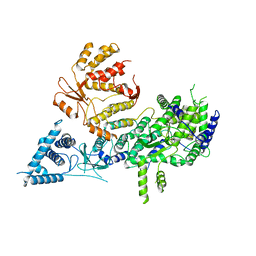 | | Apo-structure of Lassa virus L protein (well-resolved polymerase core) [APO-CORE] | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7OEB
 
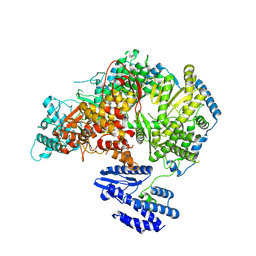 | | Lassa virus L protein bound to 3' promoter RNA (well-resolved endonuclease) [3END-ENDO] | | Descriptor: | 3' vRNA, MAGNESIUM ION, RNA-directed RNA polymerase L, ... | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-02 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
8W8E
 
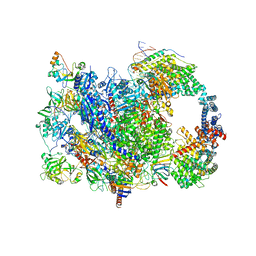 | | human co-transcriptional RNA capping enzyme RNGTT | | Descriptor: | DNA (36-MER), DNA (45-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
4XLS
 
 | |
8W8F
 
 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
7QTL
 
 | | Influenza A/H7N9 polymerase elongation complex | | Descriptor: | 3' vRNA, 5' vRNA, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Cusack, S, Kouba, T. | | Deposit date: | 2022-01-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Direct observation of backtracking by influenza A and B polymerases upon consecutive incorporation of the nucleoside analog T1106.
Cell Rep, 42, 2023
|
|
7R0E
 
 | |
7R1F
 
 | |
4WTJ
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AUCC, RNA PRIMER 5'-PGG, MN2+, AND ADP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTM
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UAGG, RNA PRIMER 5'-PCC, MN2+, AND UDP | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, RNA PRIMER CC, ... | | Authors: | Edwards, T.E, Appleby, T.C, Fox III, D. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTI
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-ACGG, RNA PRIMER 5'-PCC, MN2+, AND GDP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4WTK
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-AGCC, RNA PRIMER 5'-PGG, MN2+, AND CDP | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
