6IV5
 
 | | Crystal structure of arabidopsis N6-mAMP deaminase MAPDA | | Descriptor: | Adenosine/AMP deaminase family protein, PHOSPHATE ION, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
2OOE
 
 | | Crystal structure of HAT domain of murine CstF-77 | | Descriptor: | Cleavage stimulation factor 77 kDa subunit | | Authors: | Bai, Y, Auperin, T.C, Chou, C.-Y, Chang, G.-G, Manley, J.L, Tong, L. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Murine CstF-77: Dimeric Association and Implications for Polyadenylation of mRNA Precursors.
Mol.Cell, 25, 2007
|
|
2OND
 
 | | Crystal Structure of the HAT-C domain of murine CstF-77 | | Descriptor: | Cleavage stimulation factor 77 kDa subunit | | Authors: | Bai, Y, Auperin, T.C, Chou, C.-Y, Chang, G.-G, Manley, J.L, Tong, L. | | Deposit date: | 2007-01-23 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Murine CstF-77: Dimeric Association and Implications for Polyadenylation of mRNA Precursors.
Mol.Cell, 25, 2007
|
|
3GXQ
 
 | | Structure of ArtA and DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*AP*CP*AP*TP*G)-3'), DNA (5'-D(*AP*CP*AP*TP*GP*TP*CP*AP*TP*GP*T)-3'), Putative regulator of transfer genes ArtA | | Authors: | Ni, L, Firth, N, Schumacher, M.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes.
Nucleic Acids Res., 37, 2009
|
|
1C0O
 
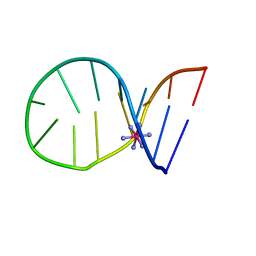 | |
3O8D
 
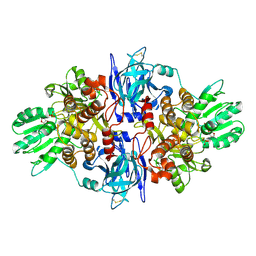 | |
4WJS
 
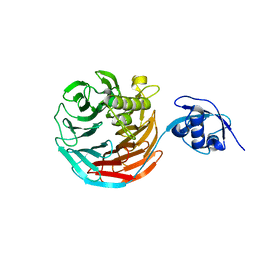 | |
3JD7
 
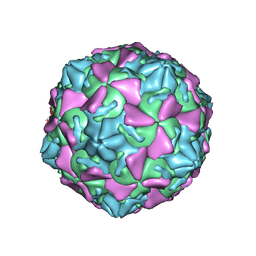 | | The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Lee, H, Shingler, K.L, Organtini, L.J, Ashley, R.E, Makhov, A.M, Conway, J.F, Hafenstein, S. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs.
Sci Adv, 2, 2016
|
|
6PUQ
 
 | |
6BID
 
 | | 1.15 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(8S,11S,14S)-11-(cyclohexylmethyl)-8-(hydroxymethyl)-5,10,13-trioxo-1,4,9,12,17,18-hexaazabicyclo[14.2.1]nonadeca-16(19),17-dien-14-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
3Q3Y
 
 | | Complex structure of HEVB EV93 main protease 3C with Compound 1 (AG7404) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, HEVB EV93 3C protease, ... | | Authors: | Costenaro, L, Kaczmarska, Z, Arnan, C, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
6PUP
 
 | |
3GDH
 
 | | Methyltransferase domain of human Trimethylguanosine Synthase 1 (TGS1) bound to m7GTP and adenosyl-homocysteine (active form) | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, R-1,2-PROPANEDIOL, S-1,2-PROPANEDIOL, ... | | Authors: | Monecke, T, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-02-24 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1.
Nucleic Acids Res., 37, 2009
|
|
6BIC
 
 | | 2.25 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6BIB
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(9S,12S,15S)-12-(cyclohexylmethyl)-9-(hydroxymethyl)-6,11,14-trioxo-1,5,10,13,18,19-hexaazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
5UKI
 
 | |
4GEK
 
 | | Crystal Structure of wild-type CmoA from E.coli | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA (cmo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bonanno, J.B, Bhosle, R, Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Nature, 498, 2013
|
|
2A0C
 
 | | Human CDK2 in complex with olomoucine II, a novel 2,6,9-trisubstituted purine cyclin-dependent kinase inhibitor | | Descriptor: | 2-{[(2-{[(1R)-1-(HYDROXYMETHYL)PROPYL]AMINO}-9-ISOPROPYL-9H-PURIN-6-YL)AMINO]METHYL}PHENOL, Cell division protein kinase 2 | | Authors: | Krystof, V, McNae, I.W, Walkinshaw, M.D, Fischer, P.M, Muller, P, Vojtesek, B, Orsag, M, Havlicek, L, Strnad, M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiproliferative activity of olomoucine II, a novel 2,6,9-trisubstituted purine cyclin-dependent kinase inhibitor
Cell.Mol.Life Sci., 62, 2005
|
|
1OZM
 
 | | Y106F mutant of Z. mobilis TGT | | Descriptor: | Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Stubbs, M.T, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Flexible adaptations in the structure of the tRNA-modifying enzyme
tRNA-guanine transglycosylase and their implications for substrate selectivity,
reaction mechanism and structure-based drug design
Chembiochem, 4, 2003
|
|
4MEM
 
 | | Crystal Structure of the rat USP11 DUSP-UBL domains | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 11 | | Authors: | Harper, S, Gratton, H.E, Cornaciu, I, Oberer, M, Scott, D.J, Emsley, J, Dreveny, I. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Catalytic Regulatory Function of Ubiquitin Specific Protease 11 N-Terminal and Ubiquitin-like Domains.
Biochemistry, 53, 2014
|
|
3J06
 
 | | CryoEM Helical Reconstruction of TMV | | Descriptor: | 5'-R(P*AP*UP*G)-3', Coat protein | | Authors: | Ge, P, Zhou, Z.H. | | Deposit date: | 2011-04-26 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1K6O
 
 | | Crystal Structure of a Ternary SAP-1/SRF/c-fos SRE DNA Complex | | Descriptor: | 5'-D(*CP*AP*CP*AP*GP*GP*AP*TP*GP*TP*CP*CP*AP*TP*AP*TP*TP*AP*GP*GP*AP*CP*A)-3', 5'-D(*TP*GP*TP*CP*CP*TP*AP*AP*TP*AP*TP*GP*GP*AP*CP*AP*TP*CP*CP*TP*GP*TP*G)-3', ETS-domain protein ELK-4, ... | | Authors: | Mo, Y, Ho, W, Johnston, K, Marmorstein, R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex.
J.Mol.Biol., 314, 2001
|
|
6G04
 
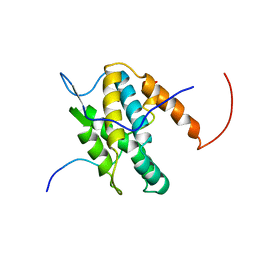 | | NMR Solution Structure of Yeast TSR2(1-152) in Complex with S26A(100-119) | | Descriptor: | 40S ribosomal protein S26-A, Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
1XUX
 
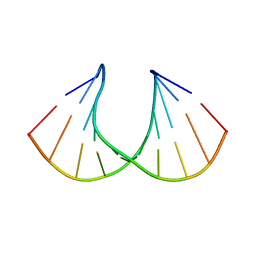 | | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogs | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(NMS)P*AP*CP*GP*C)-3') | | Authors: | Pattanayek, R, Sethaphong, L, Pan, C, Prhavc, M, Prakash, T.P, Manoharan, M, Egli, M. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural rationalization of a large difference in RNA affinity despite a small difference in chemistry between two 2'-O-modified nucleic acid analogues.
J.Am.Chem.Soc., 126, 2004
|
|
