8P3A
 
 | | bacteriophage T5 l-alanoyl-d-glutamate peptidase Zn2+/Ca2+ form | | Descriptor: | CALCIUM ION, L-alanyl-D-glutamate peptidase, ZINC ION | | Authors: | Prokhorov, D.A, Kutyshenko, V.P, Mikoulinskaia, G.V. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-01-15 | | Method: | SOLUTION NMR | | Cite: | Conservative Tryptophan Residue in the Vicinity of an Active Site of the M15 Family l,d-Peptidases: A Key Element in the Catalysis.
Int J Mol Sci, 24, 2023
|
|
4WAM
 
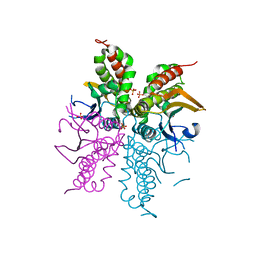 | |
4WB4
 
 | | wt SA11 NSP4_CCD | | Descriptor: | CALCIUM ION, Non-structural glycoprotein NSP4 | | Authors: | Viskovska, M, Sastri, N.P, Hyser, J.M, Tanner, M.R, Horton, L.B, Sankaran, B, Prasad, B.V.V, Estes, M.K. | | Deposit date: | 2014-09-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Plasticity of the Coiled-Coil Domain of Rotavirus NSP4.
J.Virol., 88, 2014
|
|
4WB9
 
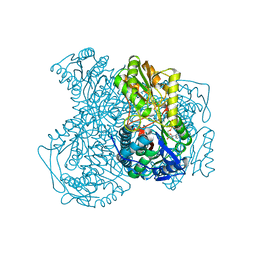 | | Human ALDH1A1 complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2014-09-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1.
Chem.Biol.Interact., 234, 2015
|
|
4WBH
 
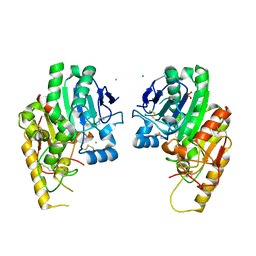 | |
4WC7
 
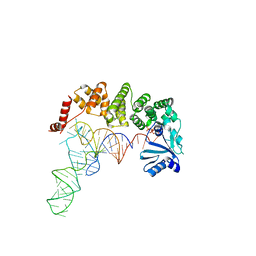 | | Structure of tRNA-processing enzyme complex 5 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Poly A polymerase, RNA (75-MER) | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2014-09-04 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Measurement of Acceptor-T Psi C Helix Length of tRNA for Terminal A76-Addition by A-Adding Enzyme.
Structure, 23, 2015
|
|
7NPU
 
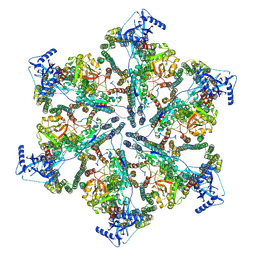 | | MycP5-free ESX-5 inner membrane complex, state I | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
4WCI
 
 | | Crystal structure of the 1st SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide (aa 378-393) from human RIN3 | | Descriptor: | CD2-associated protein, Ras and Rab interactor 3, SULFATE ION | | Authors: | Rouka, E, Simister, P.C, Janning, M, Kirsch, K.H, Krojer, T, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-04 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
4WCW
 
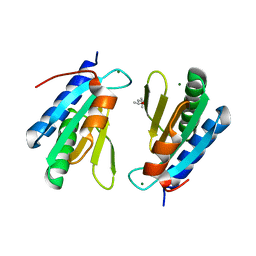 | | Ribosomal silencing factor during starvation or stationary phase (RsfS) from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, Ribosomal silencing factor RsfS | | Authors: | Li, X, Sun, Q, Jiang, C, Yang, K, Hung, L, Zhang, J, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Ribosomal Silencing Factor Bound to Mycobacterium tuberculosis Ribosome.
Structure, 23, 2015
|
|
4WD0
 
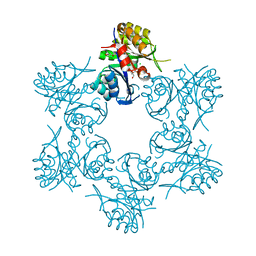 | | Crystal structure of HisAp form Arthrobacter aurescens | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | MICHALSKA, K, VERDUZCO-CASTRO, E.A, ENDRES, M, BARONA-GOMEZ, F, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of HisAp form Arthrobacter aurescens
To Be Published
|
|
4WDA
 
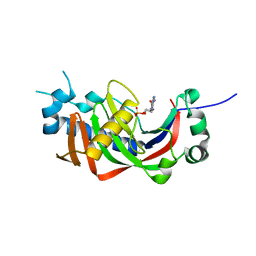 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation P296G, complexed with 2'-AMP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WDI
 
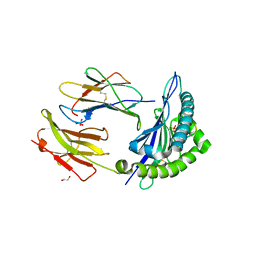 | |
7NPR
 
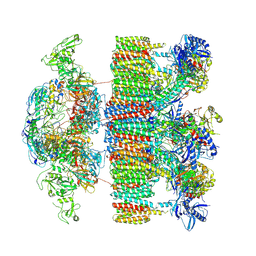 | | Structure of an intact ESX-5 inner membrane complex, Composite C3 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
4V6S
 
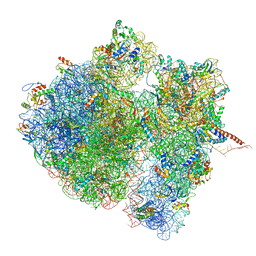 | | Structural characterization of mRNA-tRNA translocation intermediates (class 3 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (13.1 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4WE8
 
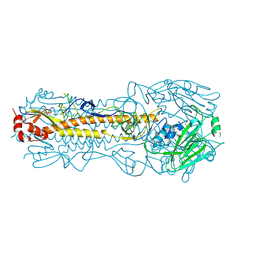 | | The crystal structure of hemagglutinin of influenza virus A/Victoria/361/2011 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
7NPV
 
 | | MycP5-free ESX-5 inner membrane complex, State II | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
4WEF
 
 | | Structure of the Hemagglutinin-neuraminidase from Human parainfluenza virus type III: complex with difluorosialic acid | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, (3R,4R,5R,6R)-5-(acetylamino)-3-fluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-3,4,5,6-tetrahydropyranium-2-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Pilling, P, Barrett, S, McKimm-Breschkin, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN)
Antiviral Res., 123, 2015
|
|
4WC9
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation F235L | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4WCK
 
 | |
4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | Descriptor: | L-amino acid ligase | | Authors: | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4WDC
 
 | |
4WDR
 
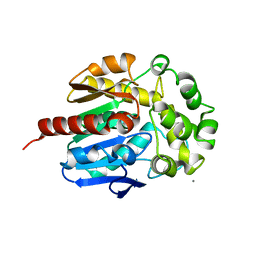 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta-Smatanova, I. | | Deposit date: | 2014-09-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB mutant (L177W) from Sphingobium japonicum UT26
Acs Catalysis, 2016
|
|
4WE5
 
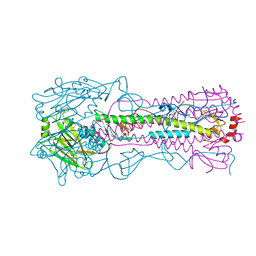 | | The crystal structure of hemagglutinin from A/Port Chalmers/1/1973 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
4WEG
 
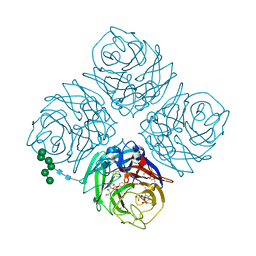 | | influenza virus neuraminidase N9 in complex 2,3-difluorosialic acid | | Descriptor: | (2R,3R,4R,5R,6R)-5-acetamido-2,3-difluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]tetrahydro-2H-pyran-2-carboxylic acid, (3R,4R,5R,6R)-5-(acetylamino)-3-fluoro-4-hydroxy-6-[(1R,2R)-1,2,3-trihydroxypropyl]-3,4,5,6-tetrahydropyranium-2-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Pilling, P, Barrett, S, McKimm-Breschkin, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic mechanism and novel receptor binding sites of human parainfluenza virus type 3 hemagglutinin-neuraminidase (hPIV3 HN)
Antiviral Res., 123, 2015
|
|
4WEK
 
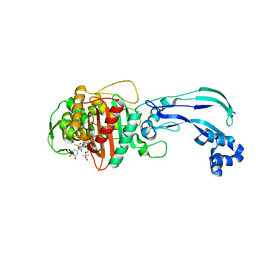 | | Crystal structure of Pseudomonas aeruginosa PBP3 with a R4 substituted vinyl monocarbam | | Descriptor: | (3S,4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-3-ethenyl-4-formyl-1-[({3-(5-hydroxy-4-oxo-3,4-dihydropyridin-2-yl)-4-[3-(methylsulfonyl)propyl]-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl}sulfonyl)amino]-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oic acid, Penicillin-binding protein 3 | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | SAR and Structural Analysis of Siderophore-Conjugated Monocarbam Inhibitors of Pseudomonas aeruginosa PBP3.
Acs Med.Chem.Lett., 6, 2015
|
|
