6VJ7
 
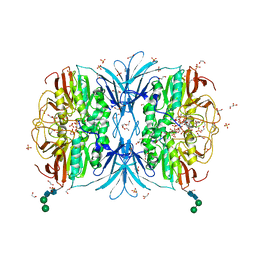 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine 5'-(beta,gamma imido)triphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
1H80
 
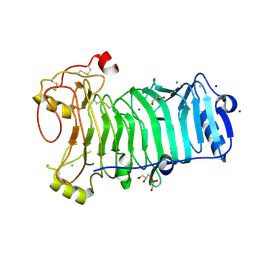 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE- 3,6-ANHYDRO-D-GALACTOSE-2-SULFATE 4 GALACTOHYDROLASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2001-01-22 | | Release date: | 2001-11-27 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Iota-Carrageenase of Alteromonas Fortis. A Beta-Helix Fold-Containing Enzyme for the Degradation of a Highly Polyanionic Polysaccharide
J.Biol.Chem., 276, 2001
|
|
6W7P
 
 | |
6W2M
 
 | |
6VUE
 
 | | wild-type choline TMA lyase in complex with 1-methyl-1,2,3,6-tetrahydropyridin-3-ol | | Descriptor: | (3S)-1-methyl-1,2,3,6-tetrahydropyridin-3-ol, Choline trimethylamine-lyase, SODIUM ION | | Authors: | Ortega, M.A, Drennan, C.L. | | Deposit date: | 2020-02-15 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of a Cyclic Choline Analog That Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria.
Acs Med.Chem.Lett., 11, 2020
|
|
6W4U
 
 | |
6MX2
 
 | |
6VUR
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT366 | | Descriptor: | 2-({[(3S)-1-methylpiperidin-3-yl]methyl}sulfanyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-amine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6W26
 
 | | Terpenoid Cyclase FgGS in Complex with Mg, Inorganic Pyrophosphate, and Imidazole | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of the cryptic function of terpene cyclases as aromatic prenyltransferases.
Nat Commun, 11, 2020
|
|
6W3L
 
 | | APE1 exonuclease substrate complex wild-type | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
4WZV
 
 | | Crystal structure of a hydroxamate based inhibitor EN140 in complex with the MMP-9 catalytic domain | | Descriptor: | (2R)-4-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-N-hydroxy-2-{[(4'-methoxybiphenyl-4-yl)sulfonyl](propan-2-yloxy)amino}butanamide, 1,2-ETHANEDIOL, AZIDE ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2014-11-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
6FAW
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2c-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-[2-(2-methoxyethoxy)phenyl]-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FGB
 
 | | Human FcRn extra-cellular domain complexed with Fab fragment of Rozanolixizumab | | Descriptor: | 1519.g57- Heavy chain, 1519.g57- Light chain, Beta-2-microglobulin, ... | | Authors: | Sarkar, K, Ceska, T, Meier, C. | | Deposit date: | 2018-01-10 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Generation and characterization of a high affinity anti-human FcRn antibody, rozanolixizumab, and the effects of different molecular formats on the reduction of plasma IgG concentration.
MAbs, 10, 2018
|
|
6FHX
 
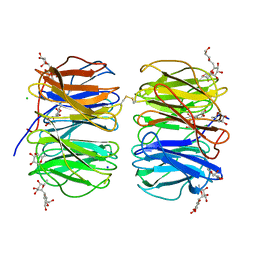 | | Photorhabdus asymbiotica lectin (PHL) in complex with synthetic C-fucoside | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{S})-2-[[(2~{R},3~{S},4~{R},6~{R})-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]methyl]-6-methyl-oxane-3,4,5-triol, CHLORIDE ION, Lectin PHL, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Fucosylated inhibitors of recently identified bangle lectin from Photorhabdus asymbiotica.
Sci Rep, 9, 2019
|
|
6FMP
 
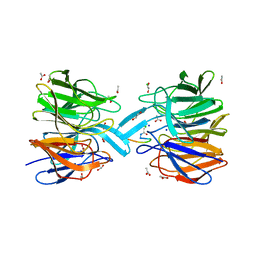 | | Keap1 - peptide complex | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ACY-ASP-GLU-GLU-THR-GLY-GLU-PHE, ... | | Authors: | Talapatra, S.K, Kozielski, F, Wells, G, Georgakopoulos, N.D. | | Deposit date: | 2018-02-01 | | Release date: | 2018-08-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Modified Peptide Inhibitors of the Keap1-Nrf2 Protein-Protein Interaction Incorporating Unnatural Amino Acids.
Chembiochem, 19, 2018
|
|
6FQV
 
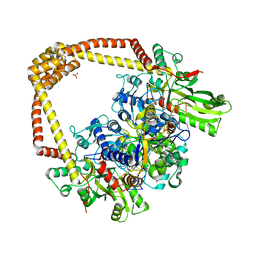 | | 2.60A BINARY COMPLEX OF S.AUREUS GYRASE with UNCLEAVED DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, DNA gyrase subunit B,DNA gyrase subunit B, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
6L56
 
 | | Fe(II) loaded Tegillarca granosa ferritin | | Descriptor: | FE (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85300577 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
6KZY
 
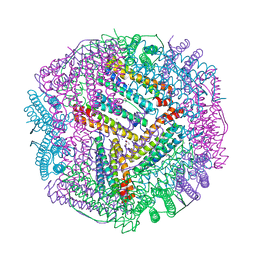 | | Cu(II) loaded Tegillarca granosa ferritin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, Ferritin, ... | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-09-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.30057073 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
6YL5
 
 | | Crystal structure of the SAM-SAH riboswitch with SAH | | Descriptor: | Chains: A,B,C,D,E,F,G,H,I,J,K,L, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6LVS
 
 | |
4MB5
 
 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4US5
 
 | | Crystal Structure of apo-MsnO8 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maier, S, Pflueger, T, Loesgen, S, Asmus, K, Broetz, E, Paululat, T, Zeeck, A, Andrade, S, Bechthold, A. | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into the Bioactivity of Mensacarcin and Epoxide Formation by Msno8.
Chembiochem, 15, 2014
|
|
4M7P
 
 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
3J7T
 
 | | Calcium atpase structure with two bound calcium ions determined by electron crystallography of thin 3D crystals | | Descriptor: | CALCIUM ION, SODIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Yonekura, K, Kato, K, Ogasawara, M, Tomita, M, Toyoshima, C. | | Deposit date: | 2014-08-07 | | Release date: | 2015-02-18 | | Last modified: | 2016-09-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.4 Å) | | Cite: | Electron crystallography of ultrathin 3D protein crystals: atomic model with charges
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3K2L
 
 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) | | Descriptor: | CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2, SODIUM ION, ... | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Soundararajan, M, Krojer, T, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of Down Syndrome Kinases, DYRKs, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
