1V0N
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
3ZHY
 
 | | Structure of Mycobacterium tuberculosis DXR in complex with a di- substituted fosmidomycin analogue | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bjorkelid, C, Jansson, A.M, Bergfors, T, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2012-12-30 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dxr Inhibition by Potent Mono- and Disubstituted Fosmidomycin Analogues.
J.Med.Chem., 56, 2013
|
|
1P4S
 
 | |
3C8Z
 
 | | The 1.6 A Crystal Structure of MshC: The Rate Limiting Enzyme in the Mycothiol Biosynthetic Pathway | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Cysteinyl-tRNA synthetase, ... | | Authors: | Tremblay, L.W, Fan, F, Vetting, M.W, Blanchard, J.S. | | Deposit date: | 2008-02-14 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of Mycobacterium smegmatis MshC: the penultimate enzyme in the mycothiol biosynthetic pathway.
Biochemistry, 47, 2008
|
|
1F8I
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE LYASE:NITROPROPIONATE:GLYOXYLATE COMPLEX FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | GLYOXYLIC ACID, ISOCITRATE LYASE, MAGNESIUM ION, ... | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-30 | | Release date: | 2000-12-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1NWA
 
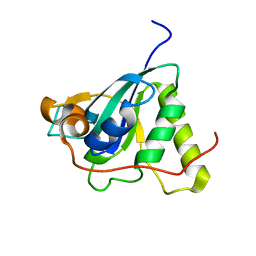 | | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Taylor, A.B, Benglis Jr, D.M, Dhandayuthapani, S, Hart, P.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine
J.Bacteriol., 185, 2003
|
|
1F8M
 
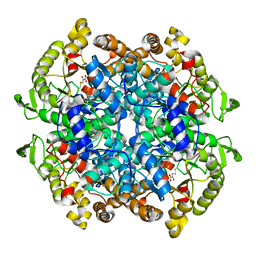 | | CRYSTAL STRUCTURE OF 3-BROMOPYRUVATE MODIFIED ISOCITRATE LYASE (ICL) FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ISOCITRATE LYASE, MAGNESIUM ION, PYRUVIC ACID | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-30 | | Release date: | 2000-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
4A03
 
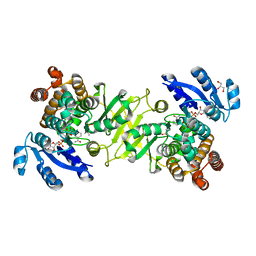 | | Crystal Structure of Mycobacterium tuberculosis DXR in complex with the antibiotic FR900098 and cofactor NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[ethanoyl(hydroxy)amino]propylphosphonic acid, GLYCEROL, ... | | Authors: | Bjorkelid, C, Bergfors, T, Jones, T.A. | | Deposit date: | 2011-09-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Studies on Mycobacterium Tuberculosis Dxr in Complex with the Antibiotic Fr-900098.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2BYO
 
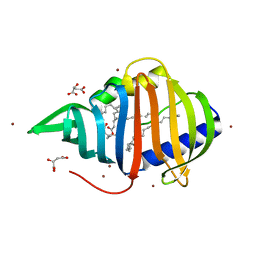 | | Crystal structure of Mycobacterium tuberculosis lipoprotein LppX (Rv2945c) | | Descriptor: | ACETATE ION, ALPHA-LINOLENIC ACID, D-MALATE, ... | | Authors: | Sulzenbacher, G, Canaan, S, Roig-Zamboni, V, Maurin, D, Gicquel, B, Bourne, Y. | | Deposit date: | 2005-08-03 | | Release date: | 2006-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Lppx is a Lipoprotein Required for the Translocation of Phthiocerol Dimycocerosates to the Surface of Mycobacterium Tuberculosis.
Embo J., 25, 2006
|
|
3DWG
 
 | | Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis | | Descriptor: | 9.5 kDa culture filtrate antigen cfp10A, Cysteine synthase B, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jurgenson, C.T, Burns, K.E, Begley, T.P, Ealick, S.E. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of a sulfur carrier protein complex found in the cysteine biosynthetic pathway of Mycobacterium tuberculosis.
Biochemistry, 47, 2008
|
|
9AZ5
 
 | | Mycolicibacterium smegmatis ClpS with bound Mg2+ | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Presloid, C.J, Jiang, J, Beardslee, P.C, Anderson, H.R, Swayne, T.M, Schmitz, K.R. | | Deposit date: | 2024-03-10 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | ClpS Directs Degradation of N-Degron Substrates With Primary Destabilizing Residues in Mycolicibacterium smegmatis.
Mol.Microbiol., 123, 2025
|
|
9AYO
 
 | | Mycolicibacterium smegmatis ClpS with bound PheAla and Ni2+ | | Descriptor: | ALANINE, ATP-dependent Clp protease adapter protein ClpS, MAGNESIUM ION, ... | | Authors: | Presloid, C.J, Jiang, J, Beardslee, P.C, Anderson, H.R, Swayne, T.M, Schmitz, K.R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ClpS Directs Degradation of N-Degron Substrates With Primary Destabilizing Residues in Mycolicibacterium smegmatis.
Mol.Microbiol., 123, 2025
|
|
9B06
 
 | | Mycolicibacterium smegmatis ClpS with LeuThr dipeptide and Mg2+ | | Descriptor: | (2S)-2-amino-4-methylpentanal, ATP-dependent Clp protease adapter protein ClpS, MAGNESIUM ION, ... | | Authors: | Presloid, C.J, Jiang, J, Beardslee, P.C, Anderson, H.R, Swayne, T.M, Schmitz, K.R. | | Deposit date: | 2024-03-11 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ClpS Directs Degradation of N-Degron Substrates With Primary Destabilizing Residues in Mycolicibacterium smegmatis.
Mol.Microbiol., 123, 2025
|
|
9AYP
 
 | | Mycolicibacterium smegmatis ClpS with bound Co2+ | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS, COBALT (II) ION, MAGNESIUM ION | | Authors: | Presloid, C.J, Jiang, J, Beardslee, P.C, Anderson, H.R, Swayne, T.M, Schmitz, K.R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | ClpS Directs Degradation of N-Degron Substrates With Primary Destabilizing Residues in Mycolicibacterium smegmatis.
Mol.Microbiol., 123, 2025
|
|
9AYN
 
 | | Mycolicibacterium smegmatis ClpS with bound Ni2+ | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS, MAGNESIUM ION, NICKEL (II) ION | | Authors: | Presloid, C.J, Jiang, J, Beardslee, P.C, Anderson, H.R, Swayne, T.M, Schmitz, K.R. | | Deposit date: | 2024-03-08 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | ClpS Directs Degradation of N-Degron Substrates With Primary Destabilizing Residues in Mycolicibacterium smegmatis.
Mol.Microbiol., 123, 2025
|
|
1UZM
 
 | | MabA from Mycobacterium tuberculosis | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER PROTEIN] REDUCTASE, CESIUM ION | | Authors: | Cohen-Gonsaud, M, Ducasse, S, Quemard, A, Labesse, G. | | Deposit date: | 2004-03-14 | | Release date: | 2005-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of Maba from Mycobacterium Tuberculosis, a Reductase Involved in Long-Chain Fatty Acid Biosynthesis.
J.Mol.Biol., 320, 2002
|
|
1UZL
 
 | | MabA from Mycobacterium tuberculosis | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER PROTEIN] REDUCTASE, CESIUM ION | | Authors: | Cohen-Gonsaud, M, Ducasse, S, Quemard, A, Labesse, G. | | Deposit date: | 2004-03-14 | | Release date: | 2005-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Maba from Mycobacterium Tuberculosis, a Reductase Involved in Long-Chain Fatty Acid Biosynthesis.
J.Mol.Biol., 320, 2002
|
|
1N4G
 
 | | Structure of CYP121, a Mycobacterial P450, in Complex with Iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D, Mowat, C.G, McLean, K.J, Richmond, A, Chapman, S.K, Walkinshaw, M.D, Munro, A.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-31 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450.
J.Biol.Chem., 278, 2003
|
|
1F61
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE LYASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ISOCITRATE LYASE, MAGNESIUM ION | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K.H, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-19 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1N40
 
 | | Atomic structure of CYP121, a mycobacterial P450 | | Descriptor: | Cytochrome P450 121, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Leys, D, Mowat, C.G, McLean, K.J, Richmond, A, Chapman, S.K, Walkinshaw, M.D, Munro, A.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 A reveals novel features of cytochrome P450.
J.Biol.Chem., 278, 2003
|
|
1V0L
 
 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1RJN
 
 | | The Crystal Structure of MenB (Rv0548c) from Mycobacterium tuberculosis in Complex with the CoA Portion of Naphthoyl CoA | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, COENZYME A, menB | | Authors: | Johnston, J.M, Arcus, V.L, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-19 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of naphthoate synthase (MenB) from Mycobacterium tuberculosis in both native and product-bound forms.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1RJM
 
 | | Crystal Structure of MenB (Rv0548c) from Mycobacterium tuberculosis | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, MenB | | Authors: | Johnston, J.M, Arcus, V.L, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-19 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of naphthoate synthase (MenB) from Mycobacterium tuberculosis in both native and product-bound forms.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6S2B
 
 | | Structure of beta-fructofuranosidase from Schwanniomyces occidentalis complexed with fructosyl-erythritol | | Descriptor: | (2~{S},3~{R})-4-[(2~{R},3~{S},4~{S},5~{R})-2,5-bis(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxybutane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-06-20 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | New insights into the molecular mechanism behind mannitol and erythritol fructosylation by beta-fructofuranosidase from Schwanniomyces occidentalis.
Sci Rep, 11, 2021
|
|
6S1T
 
 | |
