5XDW
 
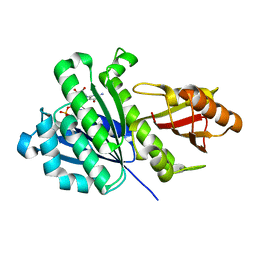 | | Staphylococcus aureus FtsZ 12-316 G196S | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
4XVX
 
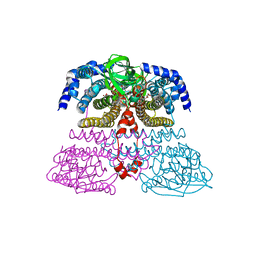 | | Crystal structure of an acyl-ACP dehydrogenase | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acyl-[acyl-carrier-protein] dehydrogenase MbtN, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Chai, A, Johnston, J.M, Bunker, R.D, Lott, J.S, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A covalent adduct of MbtN, an acyl-ACP dehydrogenase from Mycobacterium tuberculosis, reveals an unusual acyl-binding pocket.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5MCE
 
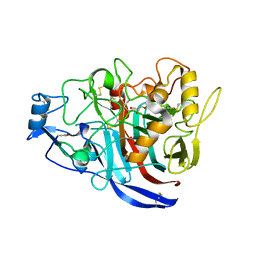 | | Radiation damage to GH7 Family Cellobiohydrolase from Daphnia pulex: Dose (DWD) 5.43 MGy | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL, SULFATE ION | | Authors: | Bury, C.S, McGeehan, J.E, Ebrahim, A, Garman, E.F. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | OH cleavage from tyrosine: debunking a myth.
J Synchrotron Radiat, 24, 2017
|
|
8PRN
 
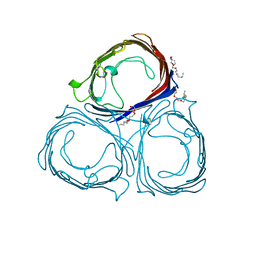 | | E1M, K50A, R52A, D97A, E99A MUTANT OF RH. BLASTICA PORIN | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, PORIN | | Authors: | Maveyraud, L, Schmid, B, Schulz, G.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Porin mutants with new channel properties.
Protein Sci., 7, 1998
|
|
1T4P
 
 | | Arginase-dehydro-ABH complex | | Descriptor: | Arginase 1, MANGANESE (II) ION, [(1E,5S)-5-AMINO-5-CARBOXYPENT-1-ENYL](TRIHYDROXY)BORATE(1-) | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Han, S, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
6RV4
 
 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
3NCF
 
 | | A mutant human Prolactin receptor antagonist H30A in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
1T5Z
 
 | | Crystal Structure of the Androgen Receptor Ligand Binding Domain (LBD) with DHT and a peptide derived from its physiological coactivator ARA70 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 4 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
3A6P
 
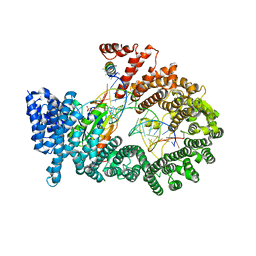 | | Crystal structure of Exportin-5:RanGTP:pre-miRNA complex | | Descriptor: | 13-mer peptide, Exportin-5, GTP-binding nuclear protein Ran, ... | | Authors: | Okada, C, Yamashita, E, Lee, S.J, Shibata, S, Katahira, J, Nakagawa, A, Yoneda, Y, Tsukihara, T. | | Deposit date: | 2009-09-07 | | Release date: | 2009-12-08 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | A high-resolution structure of the pre-microRNA nuclear export machinery
Science, 326, 2009
|
|
1WV1
 
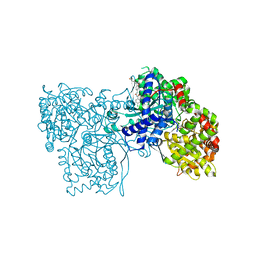 | | Crystallographic studies on acyl ureas, a new class of inhibitors of glycogenphosphorylase. Broad specificity of the allosteric site | | Descriptor: | 5-[3-({[(2,4-DICHLOROBENZOYL)AMINO]CARBONYL}AMINO)-2-METHYLPHENOXY]PENTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M.N, Chrysina, E.D, Leonidas, D.D, Klabunde, T, Wendt, K.U, Defossa, E. | | Deposit date: | 2004-12-10 | | Release date: | 2005-12-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
1T63
 
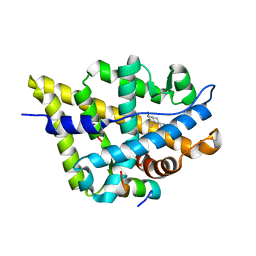 | | Crystal Structure of the Androgen Receptor Ligand Binding Domain with DHT and a peptide derived from its physiological coactivator GRIP1 NR box3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 2 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
7YWP
 
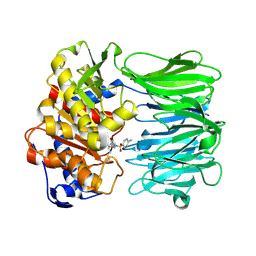 | | Closed conformation of Oligopeptidase B from Serratia proteomaculans with covalently bound TCK | | Descriptor: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Inhibitor-Bound Bacterial Oligopeptidase B in the Closed State: Similarity and Difference between Protozoan and Bacterial Enzymes.
Int J Mol Sci, 24, 2023
|
|
6TV3
 
 | | HumRadA1 in complex with 3-amino-2-naphthoic acid | | Descriptor: | 3-azanylnaphthalene-2-carboxylic acid, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Marsh, M.E, Hyvonen, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
2HIO
 
 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN | | Descriptor: | PROTEIN (HISTONE H2A), PROTEIN (HISTONE H2B), PROTEIN (HISTONE H3), ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1999-06-15 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
2HLD
 
 | | Crystal structure of yeast mitochondrial F1-ATPase | | Descriptor: | ATP synthase alpha chain, mitochondrial, ATP synthase beta chain, ... | | Authors: | Kabaleeswaran, V, Puri, N, Walker, J.E, Leslie, A.G, Mueller, D.M. | | Deposit date: | 2006-07-06 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel features of the rotary catalytic mechanism revealed in the structure of yeast F(1) ATPase.
Embo J., 25, 2006
|
|
7ABR
 
 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
1TBJ
 
 | | H141A mutant of rat liver arginase I | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-20 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
5MCJ
 
 | | Radiation damage to GH7 Family Cellobiohydrolase from Daphnia pulex: Dose (DWD) 14.1 MGy | | Descriptor: | Cellobiohydrolase CHBI, GLYCEROL, SULFATE ION | | Authors: | Bury, C.S, McGeehan, J.E, Ebrahim, A, Garman, E.F. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | OH cleavage from tyrosine: debunking a myth.
J Synchrotron Radiat, 24, 2017
|
|
7ADQ
 
 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno-Chicano, T.M, Ebrahim, A.E, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | Deposit date: | 2020-09-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
6UJX
 
 | | HIV-1 wild-type reverse transcriptase-DNA complex with (-)-FTC-TP | | Descriptor: | DNA primer, DNA template, MAGNESIUM ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2019-10-03 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.70451212 Å) | | Cite: | Elucidating molecular interactions ofL-nucleotides with HIV-1 reverse transcriptase and mechanism of M184V-caused drug resistance.
Commun Biol, 2, 2019
|
|
8PN6
 
 | | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus | | Descriptor: | Genome polyprotein, Serine protease subunit NS2B | | Authors: | Ni, X, Fairhead, M, Balcomb, B.H, Aschenbrenner, J.C, Ferreira, L.M, Godoy, A.S, Lithgo, R.M, MacLean, E.M, Marples, P.G, Thompson, W, Tomlinson, C.W.E, Szommer, T, Wild, C, Wright, N.D, Koekemoer, L, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus
To Be Published
|
|
7SEP
 
 | |
2SNS
 
 | | STAPHYLOCOCCAL NUCLEASE. PROPOSED MECHANISM OF ACTION BASED ON STRUCTURE OF ENZYME-THYMIDINE 3(PRIME),5(PRIME)-BIPHOSPHATE-CALCIUM ION COMPLEX AT 1.5-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, THERMONUCLEASE PRECURSOR, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Legg, M.J, Cotton, F.A, Hazen Jr, E.E. | | Deposit date: | 1982-05-14 | | Release date: | 1982-07-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
Thesis, Texas Agricultural and Mechanical University, 1977
|
|
4MF3
 
 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8T42
 
 | | Model of TTLL6 MTBH1-2 bound to microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 2024
|
|
