3LVH
 
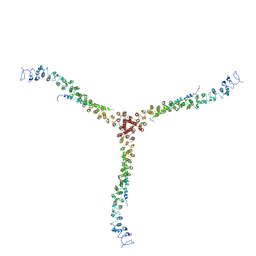 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | Deposit date: | 2010-02-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
3NPE
 
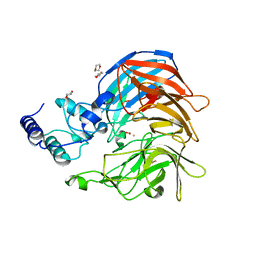 | | Structure of VP14 in complex with oxygen | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 9-cis-epoxycarotenoid dioxygenase 1, chloroplastic, ... | | Authors: | Messing, S.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-06-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid.
Plant Cell, 22, 2010
|
|
2VLG
 
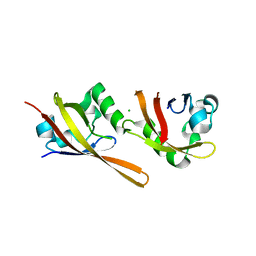 | | KinA PAS-A domain, homodimer | | Descriptor: | ACETATE ION, CHLORIDE ION, SPORULATION KINASE A | | Authors: | Lee, J, Tomchick, D.R, Brautigam, C.A, Machius, M, Kort, R, Hellingwerf, K.J, Gardner, K.H. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Changes at the Kina Pas-A Dimerization Interface Influence Histidine Kinase Function.
Biochemistry, 47, 2008
|
|
3NJO
 
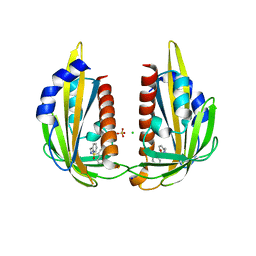 | | X-ray crystal structure of the Pyr1-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYR1, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Peterson, F.C, Volkman, B.F, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2GNW
 
 | |
7WKV
 
 | | Crystal structure of human ALKBH5 in complex with 2-oxoglutarate (2OG) and m6A-containing ssRNA | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, RNA (5'-R(P*GP*GP*(6MZ)P*C)-3'), ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2022-01-11 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
6LDT
 
 | |
5JNN
 
 | | Crystal structure of abscisic acid receptor PYL2 in complex with Phaseic acid. | | Descriptor: | (3S,4E)-5-[(1R,5R,8S)-8-hydroxy-1,5-dimethyl-3-oxo-6-oxabicyclo[3.2.1]octan-8-yl]-3-methylpent-4-enoic acid, Abscisic acid receptor PYL2 | | Authors: | Weng, J.K, Noel, J.P. | | Deposit date: | 2016-04-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-evolution of Hormone Metabolism and Signaling Networks Expands Plant Adaptive Plasticity.
Cell, 166, 2016
|
|
3NJ0
 
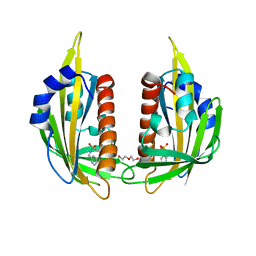 | | X-ray crystal structure of the PYL2-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJ1
 
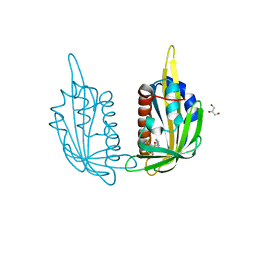 | | X-ray crystal structure of the PYL2(V114I)-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4Q2U
 
 | |
8HKC
 
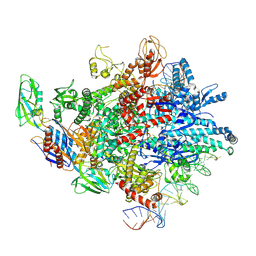 | | Cryo-EM structure of E. coli RNAP sigma32 complex | | Descriptor: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wu, S, Ma, L.X. | | Deposit date: | 2022-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural Insight into the Mechanism of sigma 32-Mediated Transcription Initiation of Bacterial RNA Polymerase.
Biomolecules, 13, 2023
|
|
7FJM
 
 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | Polyamine-transporting ATPase 13A2 | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
7FJP
 
 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
7FJQ
 
 | | Cryo EM structure of lysosomal ATPase | | Descriptor: | Polyamine-transporting ATPase 13A2, SPERMINE | | Authors: | Zhang, S.S. | | Deposit date: | 2021-08-04 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
7V4G
 
 | | Crystal structure of human ALKBH5 in complex with m6A-containing ssRNA | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RNA (5'-R(P*GP*GP*(6MZ)P*C)-3'), ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2021-08-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
4C7P
 
 | | Crystal structure of Legionella pneumophila RalF F255K mutant | | Descriptor: | GLYCEROL, RALF | | Authors: | Folly-Klan, M, Alix, E, Stalder, D, Ray, P, Duarte, L.V, Delprato, A, Zeghouf, M, Antonny, B, Campanacci, V, Roy, C.R, Cherfils, J. | | Deposit date: | 2013-09-24 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Novel Membrane Sensor Controls the Localization and Arfgef Activity of Bacterial Ralf.
Plos Pathog., 9, 2013
|
|
5YRP
 
 | | Crystal structure of the EAL domain of Mycobacterium smegmatis DcpA | | Descriptor: | MAGNESIUM ION, Sensory box/response regulator | | Authors: | Chen, H.J, li, N, Luo, Y, Jiang, Y.L, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP inMycobacterium smegmatis.
Biochem. J., 475, 2018
|
|
2JIH
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (complex-form) | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAMTS-1, CADMIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains.
J. Mol. Biol., 373, 2007
|
|
6JY1
 
 | |
3K3K
 
 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
7COW
 
 | | 353 bp di-nucleosome harboring cohesive DNA termini with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (353-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-08-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
2V4B
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (apo-form) | | Descriptor: | ADAMTS-1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Adamts-1 Reveal a Conserved Catalytic Domain and a Disintegrin-Like Domain with a Fold Homologous to Cysteine-Rich Domains.
J.Mol.Biol., 373, 2007
|
|
4ATO
 
 | | New insights into the mechanism of bacterial Type III toxin-antitoxin systems: selective toxin inhibition by a non-coding RNA pseudoknot | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, TOXI, TOXN | | Authors: | Short, F.L, Pei, X.Y, Blower, T.R, Ong, S.L, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2012-05-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity and Self-Assembly in the Control of a Bacterial Toxin by an Antitoxic Noncoding RNA Pseudoknot.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1UM8
 
 | | Crystal structure of helicobacter pylori ClpX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX | | Authors: | Kim, D.Y, Kim, K.K. | | Deposit date: | 2003-09-25 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of ClpX Molecular Chaperone from Helicobacter pylori
J.Biol.Chem., 278, 2003
|
|
