7ABN
 
 | | Structure of the reversible pyrrole-2-carboxylic acid decarboxylase PA0254/HudA | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, IMIDAZOLE, MANGANESE (II) ION, ... | | Authors: | Leys, D. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanism of Pseudomonas aeruginosa PA0254/HudA, a prFMN-Dependent Pyrrole-2-carboxylic Acid Decarboxylase Linked to Virulence.
Acs Catalysis, 11, 2021
|
|
8OX1
 
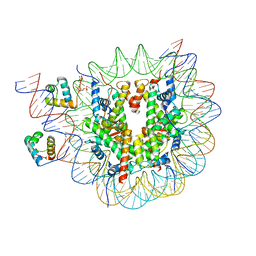 | | Structure of TRF1core in complex with telomeric nucleosome | | Descriptor: | Histone H2A type 1-C, Histone H2B type 1-C/E/F/G/I, Histone H3.1, ... | | Authors: | Hu, H, van Roon, A.M.M, Ghanim, G.E, Ahsan, B, Oluwole, A, Peak-Chew, S, Robinson, C.V, Nguyen, T.H.D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of telomeric nucleosome recognition by shelterin factor TRF1.
Sci Adv, 9, 2023
|
|
7DCE
 
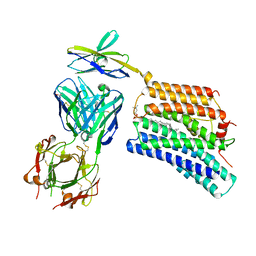 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8P4A
 
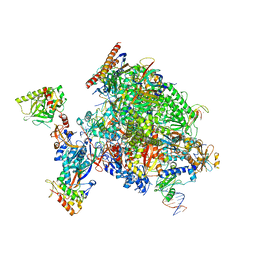 | | Structural insights into human co-transcriptional capping - structure 1 | | Descriptor: | DNA (29-MER), DNA (38-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
5DWU
 
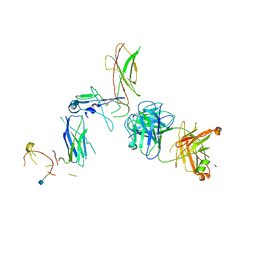 | | Beta common receptor in complex with a Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, Fab - Heavy Chain, ... | | Authors: | Dhagat, U, Parker, M.W. | | Deposit date: | 2015-09-23 | | Release date: | 2015-12-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | CSL311, a novel, potent, therapeutic monoclonal antibody for the treatment of diseases mediated by the common beta chain of the IL-3, GM-CSF and IL-5 receptors.
Mabs, 8, 2016
|
|
8P4D
 
 | | Structural insights into human co-transcriptional capping - structure 4 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P2L
 
 | |
7PKS
 
 | | Structural basis of Integrator-mediated transcription regulation | | Descriptor: | DNA Template, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Fianu, I, Chen, Y, Dienemann, C, Cramer, P. | | Deposit date: | 2021-08-26 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of Integrator-mediated transcription regulation.
Science, 374, 2021
|
|
5DW8
 
 | |
8OJM
 
 | | Galectin-3 in complex with 2,6-anhydro-3-deoxy-3-S-(beta-D-galactopyranosyl)-3-thio-D-glycero-D-galacto-heptonamide | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxane-2-carboxamide, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
5E1A
 
 | | Structure of KcsA with L24C/R117C mutations | | Descriptor: | POTASSIUM ION, antibody Fab fragment heavy chain, antibody Fab fragment light chain, ... | | Authors: | Upadhyay, V, Kim, D.M, Nimigean, C.M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational heterogeneity in closed and open states of the KcsA potassium channel in lipid bicelles.
J.Gen.Physiol., 148, 2016
|
|
8OJI
 
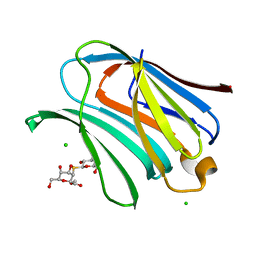 | | Galectin-3 in complex with Methyl 2,6-anhydro-3-deoxy-3-S-(b-D-galactopyranosyl)-3-thio-D-glycero-L-altro-heptonate | | Descriptor: | 1-thio-beta-D-galactopyranose, CHLORIDE ION, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
7PXH
 
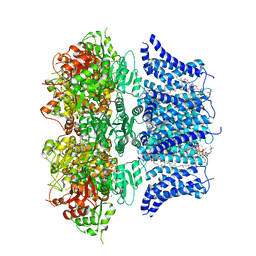 | | Emodepside-bound Drosophila Slo channel | | Descriptor: | (3~{S},6~{R},9~{S},12~{R},15~{S},18~{R},21~{S},24~{R})-4,6,10,16,18,22-hexamethyl-3,9,15,21-tetrakis(2-methylpropyl)-12,24-bis[(4-morpholin-4-ylphenyl)methyl]-1,7,13,19-tetraoxa-4,10,16,22-tetrazacyclotetracosane-2,5,8,11,14,17,20,23-octone, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, ... | | Authors: | Raisch, T, Brockmann, A, Ebbinghaus-Kintscher, U, Freigang, J, Gutbrod, O, Kubicek, J, Maertens, B, Hofnagel, O, Raunser, S. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Small molecule modulation of the Drosophila Slo channel elucidated by cryo-EM.
Nat Commun, 12, 2021
|
|
7PXG
 
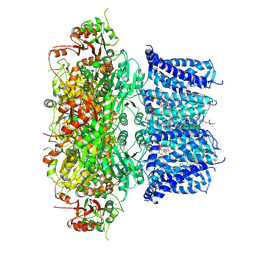 | | Verruculogen-bound Drosophila Slo channel | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Raisch, T, Brockmann, A, Ebbinghaus-Kintscher, U, Freigang, J, Gutbrod, O, Kubicek, J, Maertens, B, Hofnagel, O, Raunser, S. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Small molecule modulation of the Drosophila Slo channel elucidated by cryo-EM.
Nat Commun, 12, 2021
|
|
8OJK
 
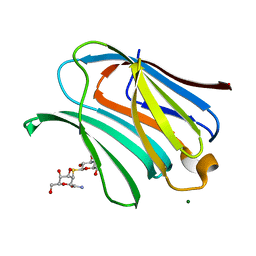 | | Galectin-3 in complex with 2,6-anhydro-3-deoxy-3-S-(beta-D-galactopyranosyl)-3-thio-D-glycero-L-altro-heptonamide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxane-2-carboxamide, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8P5Z
 
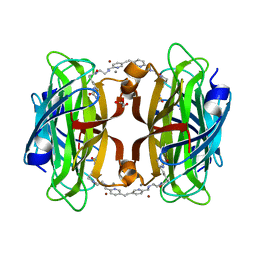 | | Artificial transfer hydrogenase with a Mn-5 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[(5-methylpyridin-2-yl)methylamino]ethyl]pentanamide, BROMIDE ION, GLYCEROL, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4G5D
 
 | |
7PXF
 
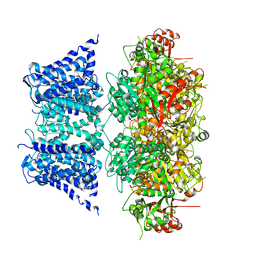 | | Ca2+ free Drosophila Slo channel | | Descriptor: | Isoform J of Calcium-activated potassium channel slowpoke, MAGNESIUM ION, POTASSIUM ION | | Authors: | Raisch, T, Brockmann, A, Ebbinghaus-Kintscher, U, Freigang, J, Gutbrod, O, Kubicek, J, Maertens, B, Hofnagel, O, Raunser, S. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Small molecule modulation of the Drosophila Slo channel elucidated by cryo-EM.
Nat Commun, 12, 2021
|
|
8P5Y
 
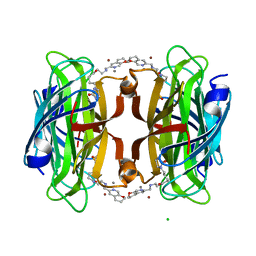 | | Artificial transfer hydrogenase with a Mn-12 cofactor and Streptavidin S112Y-K121M mutant | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)ethyl]pentanamide, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Lau, K, Wang, W, Pojer, F, Larabi, A. | | Deposit date: | 2023-05-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7PXE
 
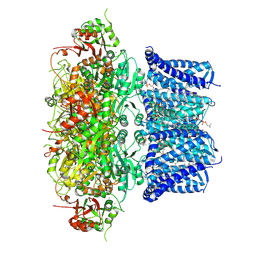 | | Ca2+ bound Drosophila Slo channel | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Raisch, T, Brockmann, A, Ebbinghaus-Kintscher, U, Freigang, J, Gutbrod, O, Kubicek, J, Maertens, B, Hofnagel, O, Raunser, S. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Small molecule modulation of the Drosophila Slo channel elucidated by cryo-EM.
Nat Commun, 12, 2021
|
|
8OJO
 
 | | Galectin-3 in complex with 2,6-Anhydro-5-S-(beta-D-galactopyranosyl)-5-thio-D-altritol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
4GAC
 
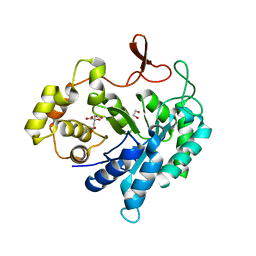 | |
7PM2
 
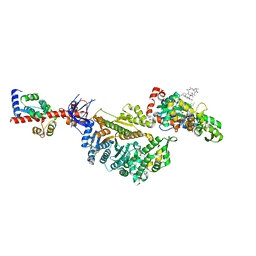 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, young JASP-stabilized F-actin, class 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PMF
 
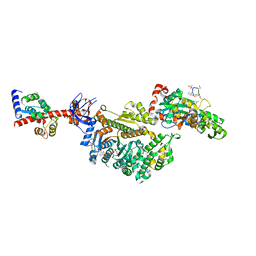 | | Cryo-EM structure of the actomyosin-V complex in the post-rigor transition state (AppNHp, central 1er, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-02 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
7PLX
 
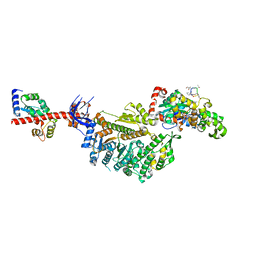 | | Cryo-EM structure of the actomyosin-V complex in the rigor state (central 1er, class 4) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Pospich, S, Sweeney, H.L, Houdusse, A, Raunser, S. | | Deposit date: | 2021-09-01 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism.
Elife, 10, 2021
|
|
