6R7N
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
8DSL
 
 | | Peptidylglycine alpha hydroxylating monooxygenase, Q272E | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
4TZ1
 
 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose | | Descriptor: | Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
6A6W
 
 | |
6KDE
 
 | |
6KDD
 
 | | endoglucanase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase, GLYCEROL, ... | | Authors: | Yu, S, Liuqing, C. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | crystal structure of an endoglucanase from Fervidobacterium pennivorans DSM9078
To Be Published
|
|
8E15
 
 | | A computationally stabilized hMPV F protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F1 protein with Fibritin peptide, F2 protein, ... | | Authors: | Huang, J, Gonzalez, K, Mousa, J, Strauch, E. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A general computational design strategy for stabilizing viral class I fusion proteins.
Biorxiv, 2023
|
|
4TX1
 
 | |
4WIL
 
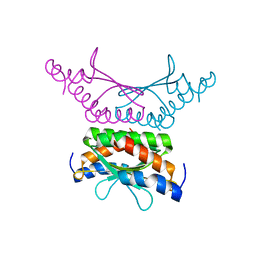 | | Crystal structure of DCoH2 S51T | | Descriptor: | Pterin-4-alpha-carbinolamine dehydratase 2 | | Authors: | Wang, D, Rose, R.B. | | Deposit date: | 2014-09-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Interactions with the Bifunctional Interface of the Transcriptional Coactivator DCoH1 Are Kinetically Regulated.
J.Biol.Chem., 290, 2015
|
|
4WJN
 
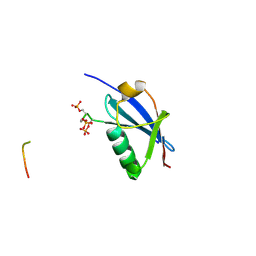 | | Crystal structure of SUMO1 in complex with phosphorylated PML | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
6KFN
 
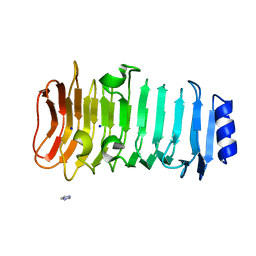 | | Crystal structure of alginate lyase from Paenibacillus sp. str. FPU-7 | | Descriptor: | IMIDAZOLE, SODIUM ION, alginate lyase | | Authors: | Itoh, T, Nakagawa, E, Yoda, M, Nakaichi, A, Hibi, T, Kimoto, H. | | Deposit date: | 2019-07-08 | | Release date: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural and biochemical characterisation of a novel alginate lyase from Paenibacillus sp. str. FPU-7.
Sci Rep, 9, 2019
|
|
4TZ5
 
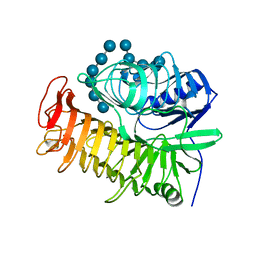 | | Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Bianchetti, C.M, Takasuka, T.E, Yik, E.J, Bergeman, L.F, Fox, B.G. | | Deposit date: | 2014-07-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active site and laminarin binding in glycoside hydrolase family 55.
J.Biol.Chem., 290, 2015
|
|
4WJP
 
 | | Crystal Structure of SUMO1 in complex with phosphorylated Daxx | | Descriptor: | Daxx, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
6KJ5
 
 | | Crystal structure of 10-Hydroxygeraniol Dehydrogenase apo form from Cantharanthus roseus | | Descriptor: | 10-hydroxygeraniol dehydrogenase, ZINC ION | | Authors: | Sandholu, A.S, Sharmila, P.M, Thulasiram, H.V, Kulkarni, K.A. | | Deposit date: | 2019-07-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structural studies on 10-hydroxygeraniol dehydrogenase: A novel linear substrate-specific dehydrogenase from Catharanthus roseus.
Proteins, 88, 2020
|
|
6KGJ
 
 | | M1Q-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Park, M.R, Kim, L, Kwon, D.H, Song, H.K. | | Deposit date: | 2019-07-11 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of the LC3B-fusion technique for biochemical and structural studies of proteins involved in the N-degron pathway.
J.Biol.Chem., 295, 2020
|
|
8DQN
 
 | |
8E14
 
 | | Cryo-EM structure of Rous sarcoma virus strand transfer complex | | Descriptor: | DNA (42-MER), DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*TP*CP*TP*TP*CP*TP*TP*TP*C)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular determinants for Rous sarcoma virus intasome assemblies involved in retroviral integration.
J.Biol.Chem., 299, 2023
|
|
1O82
 
 | | X-RAY STRUCTURE OF BACTERIOCIN AS-48 AT PH 4.5. SULPHATE BOUND FORM | | Descriptor: | GLYCEROL, PEPTIDE ANTIBIOTIC AS-48, SULFATE ION | | Authors: | Sanchez-Barrena, M.J, Martinez-Ripoll, M, Galvez, A, Martinez-Bueno, M, Maqueda, M, Cruz, V, Albert, A. | | Deposit date: | 2002-11-22 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of Bacteriocin as-48: From Soluble State to Membrane Bound State
J.Mol.Biol., 334, 2003
|
|
8DU5
 
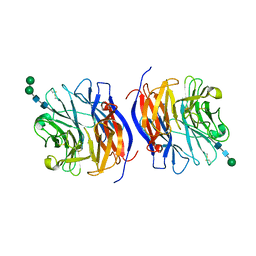 | | Murine sialidase-1 (NEU1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Sialidase-1, ... | | Authors: | Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2022-07-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the immunoregulatory sialidase NEU1.
Sci Adv, 9, 2023
|
|
4U2T
 
 | |
6KL7
 
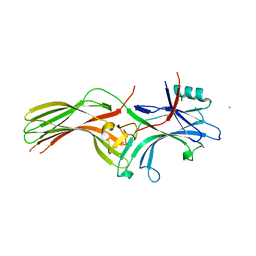 | | Beta-arrestin 1 mutant S13D/T275D | | Descriptor: | 1,2-ETHANEDIOL, BARIUM ION, Beta-arrestin-1 | | Authors: | Kang, H, Choi, H.J. | | Deposit date: | 2019-07-29 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Conformational Dynamics and Functional Implications of Phosphorylated beta-Arrestins.
Structure, 28, 2020
|
|
6KM9
 
 | | Crystal structure of SucA from Vibrio vulnificus | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Seo, P.W, Kim, J.S. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Catalysis Science And Technology, 2020
|
|
6RLV
 
 | | Trypanosoma brucei Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine | | Descriptor: | 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine, GLYCEROL, MALONATE ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
4U9Q
 
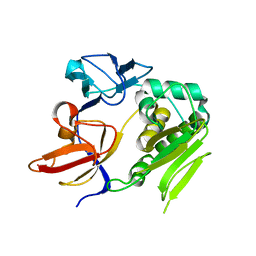 | | Crystal structure of NqrA in spacegroup P21 | | Descriptor: | Na(+)-translocating NADH-quinone reductase subunit A | | Authors: | Fritz, G. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
