7APU
 
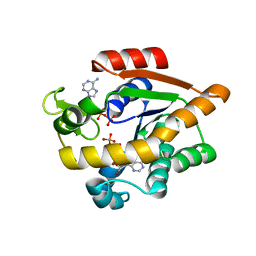 | | Structure of Adenylate kinase from Escherichia coli in complex with two ADP molecules refined at 1.36 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, SODIUM ION | | Authors: | Grundstom, C, Wolf-Watz, M, Nam, K, Sauer, U.H. | | Deposit date: | 2020-10-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Dynamic Connection between Enzymatic Catalysis and Collective Protein Motions.
Biochemistry, 60, 2021
|
|
6HDL
 
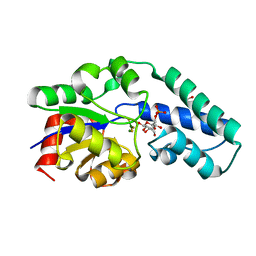 | | R49K variant of beta-phosphoglucomutase from Lactococcus lactis complexed with magnesium trifluoride and beta-G6P to 1.2 A. | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
7AW7
 
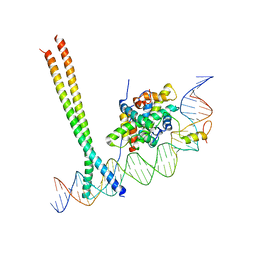 | | CCAAT-binding complex and HapX bound to Aspergillus nidulans cccA DNA | | Descriptor: | BZIP domain-containing protein, CBFD_NFYB_HMF domain-containing protein, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2020-11-06 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX.
Structure, 30, 2022
|
|
6FOA
 
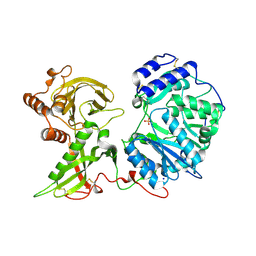 | | Human Xylosyltransferase 1 apo structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Briggs, D.C, Hohenester, E. | | Deposit date: | 2018-02-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Structural Basis for the Initiation of Glycosaminoglycan Biosynthesis by Human Xylosyltransferase 1.
Structure, 26, 2018
|
|
7B3N
 
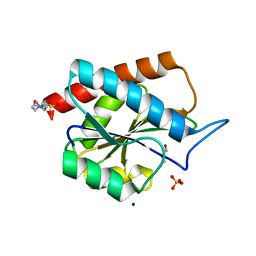 | | AmiP amidase-3 from Thermus parvatiensis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cell wall hydrolase, ... | | Authors: | Freitag-Pohl, S, Pohl, E. | | Deposit date: | 2020-12-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | AmiP from hyperthermophilic Thermus parvatiensis prophage is a thermoactive and ultrathermostable peptidoglycan lytic amidase.
Protein Sci., 32, 2023
|
|
6HDG
 
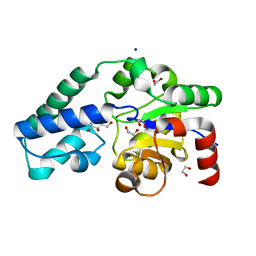 | | D170N variant of beta-phosphoglucomutase from Lactococcus lactis complexed with beta-G1P in a closed conformer to 1.2 A. | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Wood, H.P, Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6HDK
 
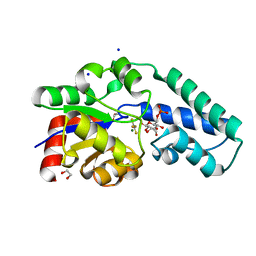 | |
7CUF
 
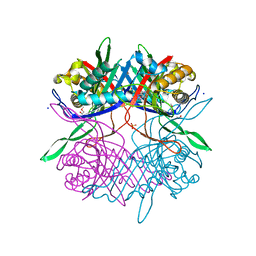 | |
7CFS
 
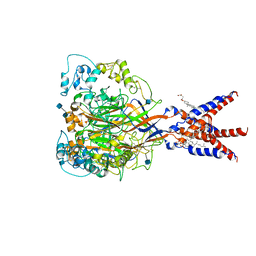 | | Cryo-EM strucutre of human acid-sensing ion channel 1a at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Sun, D.M, Liu, S.L, Li, S.Y, Yang, F, Tian, C.L. | | Deposit date: | 2020-06-28 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Elife, 9, 2020
|
|
7CM8
 
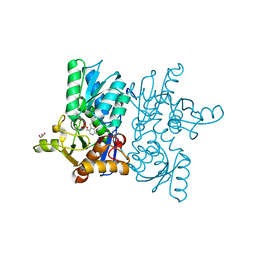 | | High resolution crystal structure of M92A mutant of O-acetyl-L-serine sulfhydrylase from Haemophilus influenzae | | Descriptor: | Cysteine synthase, GLYCEROL, SODIUM ION | | Authors: | Kaushik, A, Rahisuddin, R, Saini, N, Kumaran, S. | | Deposit date: | 2020-07-25 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mechanism of selective substrate engagement and inhibitor disengagement of cysteine synthase.
J.Biol.Chem., 296, 2020
|
|
6I03
 
 | |
7CN7
 
 | |
7CC4
 
 | |
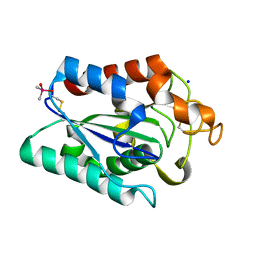
7CW1
 
 | |
6GN7
 
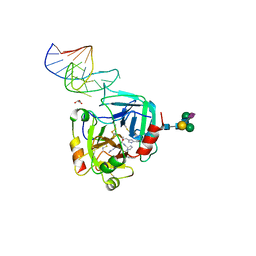 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of sodium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
6IA9
 
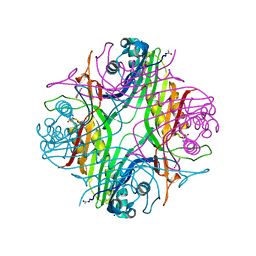 | | urate oxidase under 2000 bar (220 MPa) of argon | | Descriptor: | 8-AZAXANTHINE, ACETYL GROUP, ARGON, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
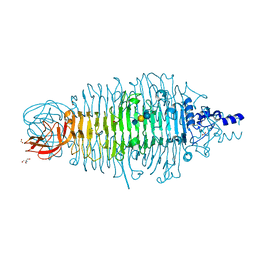
6GVR
 
 | |
7BU7
 
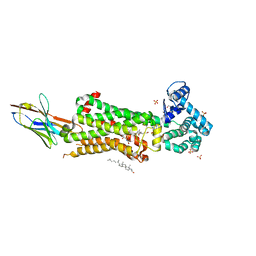 | | Structure of human beta1 adrenergic receptor bound to BI-167107 and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-04 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
7BOV
 
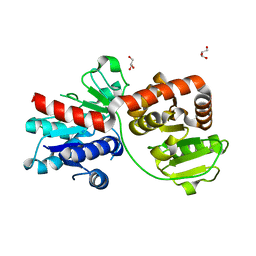 | | The Structure of Bacillus subtilis glycosyltransferase,Bs-YjiC | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Zhao, C, Liu, B, Zhao, N.L, Luo, Y.Z, Bao, R. | | Deposit date: | 2020-03-20 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and biochemical studies of the glycosyltransferase Bs-YjiC from Bacillus subtilis.
Int.J.Biol.Macromol., 166, 2021
|
|
7C6L
 
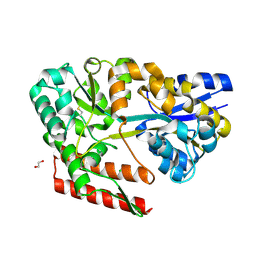 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotriose (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7ASJ
 
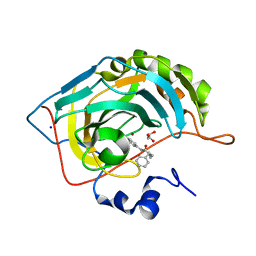 | |
7ASQ
 
 | |
7B0E
 
 | | Crystal structure of SmbA loop deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, ... | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structure of loop deletion mutant SmbA bound to a monomeric c-di-GMP
To Be Published
|
|
7B1K
 
 | | Crystal structure of phosphatidyl serine synthase (PSS) in the closed conformation with bound citrate. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CALCIUM ION, ... | | Authors: | Yildiz, O, Centola, M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of phosphatidyl serine synthase PSS reveal the catalytic mechanism of CDP-DAG alcohol O-phosphatidyl transferases
Nat Commun, 12, 2021
|
|
7B1L
 
 | | Crystal structure of phosphatidyl serine synthase (PSS) in the closed conformation with bound citrate. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, CALCIUM ION, ... | | Authors: | Yildiz, O, Centola, M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of phosphatidyl serine synthase PSS reveal the catalytic mechanism of CDP-DAG alcohol O-phosphatidyl transferases
Nat Commun, 12, 2021
|
|
