2VJD
 
 | | Torpedo Californica Acetylcholinesterase In Complex With A Non Hydrolysable Substrate Analogue, 4-Oxo-N,N,N- Trimethylpentanaminium - Orthorhombic space group - Dataset C at 150K | | Descriptor: | (4R)-4-HYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Bourgeois, D, Fournier, D, Silman, I, Sussman, J.L, Weik, M. | | Deposit date: | 2007-12-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7SQK
 
 | | Cryo-EM structure of the human augmin complex | | Descriptor: | HAUS augmin-like complex subunit 1, HAUS augmin-like complex subunit 2, HAUS augmin-like complex subunit 3, ... | | Authors: | Gabel, C.A, Chang, L. | | Deposit date: | 2021-11-05 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Molecular architecture of the augmin complex.
Nat Commun, 13, 2022
|
|
1HWK
 
 | |
2VT6
 
 | | Native Torpedo californica acetylcholinesterase collected with a cumulated dose of 9400000 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Colletier, J.P, Bourgeois, D, Sanson, B, Fournier, D, Sussman, J.L, Silman, I, Weik, M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1DHJ
 
 | |
2WG2
 
 | | NON-AGED CONJUGATE OF TORPEDO CALIFORNICA ACETYLCHOLINESTERASE WITH SOMAN (ALTERNATIVE REFINEMENT) | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (S)-METHYLPHOSPHINATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sanson, B, Nachon, F, Colletier, J.P, Froment, M.T, Toker, L, Greenblatt, H.M, Sussman, J.L, Ashani, Y, Masson, P, Silman, I, Weik, M. | | Deposit date: | 2009-04-15 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime.
J.Med.Chem., 52, 2009
|
|
2VT7
 
 | | Native Torpedo californica acetylcholinesterase collected with a cumulated dose of 800000 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Colletier, J.P, Bourgeois, D, Sanson, B, Fournier, D, Sussman, J.L, Silman, I, Weik, M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VQ6
 
 | |
8PYR
 
 | | Crystal structure of the dual T-loop phosphorylated Cdk7/CycH/Mat1 complex | | Descriptor: | 1,2-ETHANEDIOL, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Anand, K, Duster, R, Geyer, M. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of Cdk7 activation by dual T-loop phosphorylation.
Biorxiv, 2024
|
|
2WG0
 
 | | AGED CONJUGATE OF TORPEDO CALIFORNICA ACETYLCHOLINESTERASE WITH SOMAN (OBTAINED BY IN CRYSTALLO AGING) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sanson, B, Nachon, F, Colletier, J.P, Froment, M.T, Toker, L, Greenblatt, H.M, Sussman, J.L, Ashani, Y, Masson, P, Silman, I, Weik, M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime.
J.Med.Chem., 52, 2009
|
|
4EI9
 
 | | Crystal structure of Bacillus cereus TubZ, GTP-form | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Plasmid replication protein RepX | | Authors: | Hayashi, I, Hoshino, S. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid.
J.Biol.Chem., 287, 2012
|
|
4H4L
 
 | | Crystal Structure of ternary complex of HutP(HutP-L-His-Zn) | | Descriptor: | HISTIDINE, Hut operon positive regulatory protein, ZINC ION | | Authors: | Dhakshnamoorthy, B, Misono, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2012-09-17 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alternative binding modes of l-histidine guided by metal ions for the activation of the antiterminator protein HutP of Bacillus subtilis.
J.Struct.Biol., 183, 2013
|
|
1RIS
 
 | | CRYSTAL STRUCTURE OF THE RIBOSOMAL PROTEIN S6 FROM THERMUS THERMOPHILUS | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Lindahl, M, Svensson, L.A, Liljas, A, Sedelnikova, S.E, Eliseikina, I.A, Fomenkova, N.P, Nevskaya, N, Nikonov, S.V, Garber, M.B, Muranova, T.A, Rykonova, A.I, Amons, R. | | Deposit date: | 1994-05-31 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ribosomal protein S6 from Thermus thermophilus.
EMBO J., 13, 1994
|
|
1IKA
 
 | |
4HLY
 
 | |
1DHI
 
 | | LONG-RANGE STRUCTURAL EFFECTS IN A SECOND-SITE REVERTANT OF A MUTANT DIHYDROFOLATE REDUCTASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Oatley, S.J, Villafranca, J.E, Brown, K.A, Kraut, J. | | Deposit date: | 1993-10-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Long-range structural effects in a second-site revertant of a mutant dihydrofolate reductase.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1HQS
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE DEHYDROGENASE FROM BACILLUS SUBTILIS | | Descriptor: | CITRIC ACID, ISOCITRATE DEHYDROGENASE, R-1,2-PROPANEDIOL, ... | | Authors: | Singh, S.K, Matsuno, K, LaPorte, D.C, Banaszak, L.J. | | Deposit date: | 2000-12-19 | | Release date: | 2001-07-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 A. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase.
J.Biol.Chem., 276, 2001
|
|
1HWI
 
 | | COMPLEX OF THE CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH FLUVASTATIN | | Descriptor: | (3R,5S,6E)-7-[3-(4-fluorophenyl)-1-(propan-2-yl)-1H-indol-2-yl]-3,5-dihydroxyhept-6-enoic acid, ADENOSINE-5'-DIPHOSPHATE, HMG-COA REDUCTASE | | Authors: | Istvan, E.S, Deisenhofer, J. | | Deposit date: | 2001-01-09 | | Release date: | 2001-05-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mechanism for statin inhibition of HMG-CoA reductase.
Science, 292, 2001
|
|
1HJ6
 
 | | ISOCITRATE DEHYDROGENASE S113E MUTANT COMPLEXED WITH ISOPROPYLMALATE, NADP+ AND MAGNESIUM (FLASH-COOLED) | | Descriptor: | 3-ISOPROPYLMALIC ACID, GLYCEROL, ISOCITRATE DEHYDROGENASE, ... | | Authors: | Doyle, S.A, Beernink, P.T, Koshland Junior, D.E. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for a Change in Substrate Specificity: Crystal Structure of S113E Isocitrate Dehydrogenase in a Complex with Isopropylmalate, Mg2+ and Nadp
Biochemistry, 40, 2001
|
|
4E5D
 
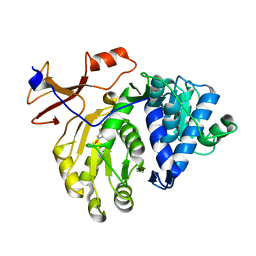 | | 2.2A resolution structure of a firefly luciferase-benzothiazole inhibitor complex | | Descriptor: | 2-(2-fluorophenyl)-6-methoxy-1,3-benzothiazole, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Throne, N, Shen, M, Auld, D.S, Inglese, J. | | Deposit date: | 2012-03-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter.
Chem.Biol., 19, 2012
|
|
4EI7
 
 | | Crystal structure of Bacillus cereus TubZ, GDP-form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Plasmid replication protein RepX | | Authors: | Hayashi, I, Hoshino, S. | | Deposit date: | 2012-04-05 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid.
J.Biol.Chem., 287, 2012
|
|
1L9A
 
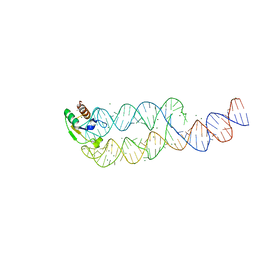 | | CRYSTAL STRUCTURE OF SRP19 IN COMPLEX WITH THE S DOMAIN OF SIGNAL RECOGNITION PARTICLE RNA | | Descriptor: | MAGNESIUM ION, METHYL MERCURY ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN, ... | | Authors: | Oubridge, C, Kuglstatter, A, Jovine, L, Nagai, K. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle.
Mol.Cell, 9, 2002
|
|
4HLX
 
 | |
1GD1
 
 | |
1GL3
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE IN COMPLEX WITH NADP AND SUBSTRATE ANALOGUE S-METHYL CYSTEINE SULFOXIDE | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE, CYSTEINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 2001-08-23 | | Release date: | 2001-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active Site Analysis of the Potential Antimicrobial Target Aspartate Semialdehyde Dehydrogenase.
Biochemistry, 40, 2001
|
|
