4B2S
 
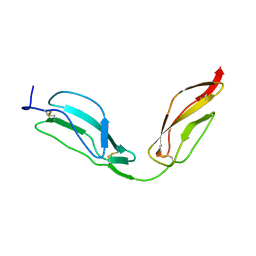 | | Solution structure of CCP modules 11-12 of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Makou, E, Mertens, H.D, Maciejewski, M, Soares, D.C, Matis, I, Schmidt, C.Q, Herbert, A.P, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ccp Modules 10-12 Illuminates Functional Architecture of the Complement Regulator, Factor H.
J.Mol.Biol., 424, 2012
|
|
8VD0
 
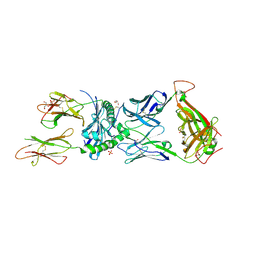 | | Human TCR ET650-4 in complex with DQ8-InsC8-15-IAPP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hybrid insulin peptide (HIP; InsC8-15-IAPP74-80),MHC class II HLA-DQ-beta-1 chimera, ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis of T cell cross-reactivity to a native and spliced self-antigens presented by HLA-DQ8
To Be Published
|
|
8OHD
 
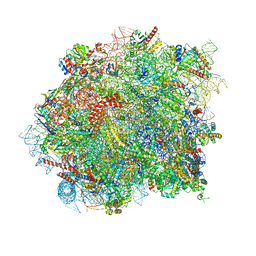 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ8
 
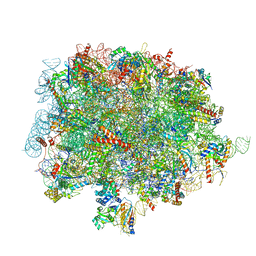 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
4BY7
 
 | | elongating RNA Polymerase II-Bye1 TLD complex | | Descriptor: | , 5'-D(*DAP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*DTP)-3', 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-18 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of RNA polymerase II complexes with Bye1, a chromatin-binding PHF3/DIDO homologue.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
8KD2
 
 | | Rpd3S in complex with 187bp nucleosome | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8OEU
 
 | | Structure of the mammalian Pol II-SPT6 complex (composite structure, Structure 4) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OF0
 
 | | Structure of the mammalian Pol II-SPT6-Elongin complex, Structure 1 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEW
 
 | | Structure of the mammalian Pol II-Elongin complex, lacking the ELOA latch (composite structure, structure 2) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEV
 
 | | Structure of the mammalian Pol II-SPT6-Elongin complex, lacking ELOA latch (composite structure, structure 3) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6Q95
 
 | |
6QDW
 
 | | Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Hodirnau, V.V, Kudlinzki, D, Mao, J, Glaubitz, C, Frangakis, A, Schwalbe, H. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cysteine oxidation and disulfide formation in the ribosomal exit tunnel.
Nat Commun, 11, 2020
|
|
160D
 
 | |
8E2Q
 
 | | Crystal structure of TadAC-1.17 in a complex with ssDNA | | Descriptor: | DNA (5'-D(P*GP*CP*GP*GP*CP*TP*(D8A)P*CP*GP*GP*A)-3'), GLYCEROL, ZINC ION, ... | | Authors: | Feliciano, P.R, Lee, S.J, Ciaramella, G. | | Deposit date: | 2022-08-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Improved cytosine base editors generated from TadA variants.
Nat.Biotechnol., 41, 2023
|
|
4BXZ
 
 | | RNA Polymerase II-Bye1 complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Kinkelin, K, Wozniak, G.G, Rothbart, S.B, Lidschreiber, M, Strahl, B.D, Cramer, P. | | Deposit date: | 2013-07-16 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Structures of RNA Polymerase II Complexes with Bye1, a Chromatin-Binding Phf3/Dido1 Homologue
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8AM9
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the elongating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-08-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
8AKN
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-07-30 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
6RYR
 
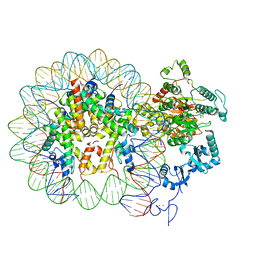 | | Nucleosome-CHD4 complex structure (single CHD4 copy) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4,Chromodomain-helicase-DNA-binding protein 4, DNA (149-MER), Histone H2A type 1, ... | | Authors: | Farnung, L, Ochmann, M, Cramer, P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nucleosome-CHD4 chromatin remodeller structure maps human disease mutations.
Elife, 9, 2020
|
|
1AA1
 
 | |
6R87
 
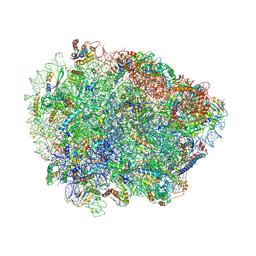 | | Yeast Vms1 (Q295L)-60S ribosomal subunit complex (pre-state without Arb1) | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
8BQD
 
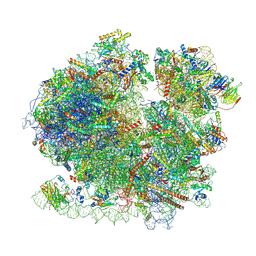 | | Yeast 80S ribosome in complex with Map1 (conformation 1) | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
1BE3
 
 | | CYTOCHROME BC1 COMPLEX FROM BOVINE | | Descriptor: | CYTOCHROME BC1 COMPLEX, FE2/S2 (INORGANIC) CLUSTER, HEME C, ... | | Authors: | Iwata, S, Lee, J.W, Okada, K, Lee, J.K, Iwata, M, Ramaswamy, S, Jap, B.K. | | Deposit date: | 1998-05-19 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex.
Science, 281, 1998
|
|
4A3G
 
 | | RNA Polymerase II initial transcribing complex with a 2nt DNA-RNA hybrid | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of initial RNA polymerase II transcription.
EMBO J., 30, 2011
|
|
8OJ0
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
7YE2
 
 | | The cryo-EM structure of C. crescentus GcrA-TACdown | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (90-MER)-non template, DNA (90-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
