7Z62
 
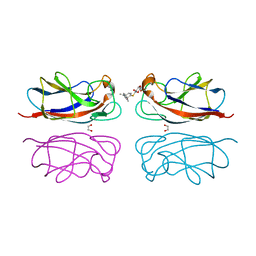 | | Structure of the LecA lectin from Pseudomonas aeruginosa in complex with a biaryl-thiogalactoside | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-[4-[1-(4-methoxyphenyl)ethenyl]phenyl]sulfanyl-oxane-3,4,5-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Varrot, A. | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of potent 1,1-diarylthiogalactoside glycomimetic inhibitors of Pseudomonas aeruginosa LecA with antibiofilm properties.
Eur.J.Med.Chem., 247, 2022
|
|
9ES9
 
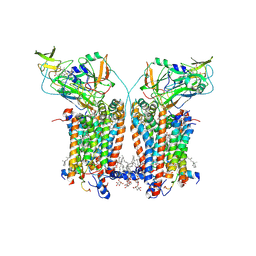 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with inhibitor DBMIB bound at plastoquinol oxidation site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 10, 2024
|
|
9JER
 
 | | Arginine decarboxylase in Aspergillus oryzae, ligand-free form | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-tryptophan decarboxylase PsiD-like domain-containing protein | | Authors: | Mikami, B, Yasukawa, K, Fujiwara, S, Takita, T, Mizutani, K, Odagaki, Y, Murakami, Y. | | Deposit date: | 2024-09-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unveiling the reaction mechanism of arginine decarboxylase in Aspergillus oryzae: Insights from crystal structure analysis.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
5MCO
 
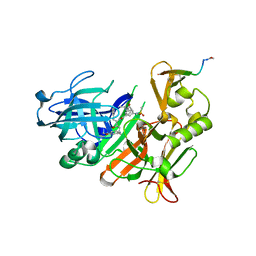 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE INHIBITOR GRL-8234 AND EXOSITE PEPTIDE | | Descriptor: | BACE-1 EXOSITE PEPTIDE, Beta-secretase 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
3KK3
 
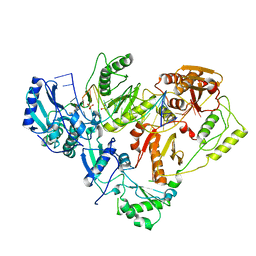 | | HIV-1 reverse transcriptase-DNA complex with GS-9148 terminated primer | | Descriptor: | 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*C*(URT))-3', 5'-D(*AP*TP*GP*GP*TP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
9F2I
 
 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with 2-amino-1,3-benzothiazol-6-ol | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-1,3-benzothiazol-6-ol, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
7QI6
 
 | | Human mitochondrial ribosome in complex with mRNA, A/P- and P/E-tRNAs at 2.98 A resolution | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Mitoribosome structure with cofactors and modifications reveals mechanism of ligand binding and interactions with L1 stalk.
Nat Commun, 15, 2024
|
|
7QI5
 
 | | Human mitochondrial ribosome in complex with mRNA, A/A-, P/P- and E/E-tRNAs at 2.63 A resolution | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Mitoribosome structure with cofactors and modifications reveals mechanism of ligand binding and interactions with L1 stalk.
Nat Commun, 15, 2024
|
|
9IZF
 
 | | Cryo-EM structure of LPA1-Gi complex with LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Kamegawa, A, Hiroaki, Y, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-08-01 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into the engagement of lysophosphatidic acid receptor 1 with different G proteins.
J.Struct.Biol., 217, 2024
|
|
7AEJ
 
 | |
8VKM
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein.
Nat Commun, 15, 2024
|
|
9IZG
 
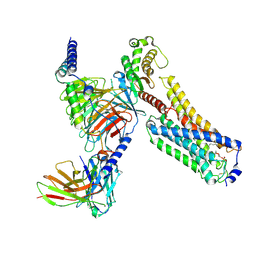 | | Cryo-EM structure of LPA1-Gq complex with LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Kmegawa, A, Hiroaki, Y, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-08-01 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural insights into the engagement of lysophosphatidic acid receptor 1 with different G proteins.
J.Struct.Biol., 217, 2024
|
|
8VKN
 
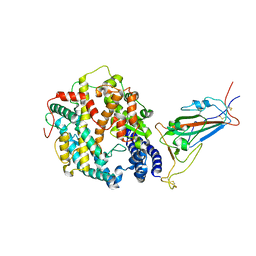 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein.
Nat Commun, 15, 2024
|
|
9EU0
 
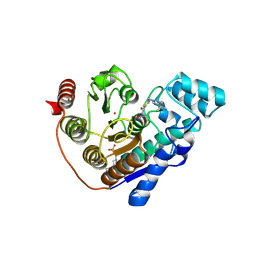 | | Crystal structure of Danio rerio HDAC6 CD2 in complex with hydrolyzed DFMO-inhibitor (ITF7209) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Bebel, A, Cellupica, E, Stevenazzi, A, Sandrone, G, Vergani, B, Caprini, G. | | Deposit date: | 2024-03-27 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanistic and Structural Insights on Difluoromethyl-1,3,4-oxadiazole Inhibitors of HDAC6.
Int J Mol Sci, 25, 2024
|
|
3L22
 
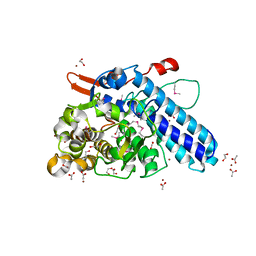 | |
9IZH
 
 | | Cryo-EM structure of LPA1-G13 complex with LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Kamegawa, A, Hiroaki, Y, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-08-01 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural insights into the engagement of lysophosphatidic acid receptor 1 with different G proteins.
J.Struct.Biol., 217, 2024
|
|
9JHA
 
 | | X-ray structure of the Haloalkane dehalogenase HaloTag7 labeled with BD626-HTL substrate | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]-2-[10,10-dimethyl-3-[(1~{R},5~{S})-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]-6-[(1~{R},5~{S})-3-oxa-8-azoniabicyclo[3.2.1]octan-8-ylidene]anthracen-9-yl]benzoic acid, CHLORIDE ION, ... | | Authors: | Zhixing, C, Kecheng, Z. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A palette of bridged bicycle-strengthened fluorophores.
Nat.Methods, 22, 2025
|
|
8VKL
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein.
Nat Commun, 15, 2024
|
|
6A1I
 
 | | Crystal structure of a synthase 1 from Santalum album | | Descriptor: | CARBONATE ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Han, X, Ko, T.P, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a synthase 1 from Santalum album in complex with ligand
To Be Published
|
|
8SVY
 
 | | MBP-Mcl1 in complex with ligand 10 | | Descriptor: | (15P)-17-chloro-33-fluoro-12-[(2-methoxyethoxy)methyl]-5,14,22-trimethyl-28-oxa-9-thia-5,6,13,14,22-pentaazaheptacyclo[27.7.1.1~4,7~.0~11,15~.0~16,21~.0~20,24~.0~30,35~]octatriaconta-1(36),4(38),6,11(15),12,16,18,20,23,29(37),30,32,34-tridecaene-23-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Macrocyclic Carbon-Linked Pyrazoles As Novel Inhibitors of MCL-1.
Acs Med.Chem.Lett., 14, 2023
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
5GLF
 
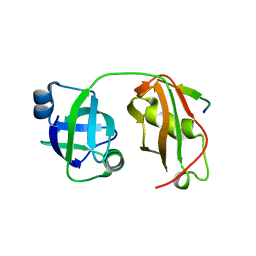 | | Structural insights into the interaction of p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease | | Descriptor: | Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Lim, J.J, Lee, Y, Yoon, S.Y, Ly, T.T, Kang, J.Y, Youn, H.-S, An, J.Y, Lee, J.-G, Park, K.R, Kim, T.G, Yang, J.K, Jun, Y, Eom, S.H. | | Deposit date: | 2016-07-11 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the interaction of human p97 N-terminal domain and SHP motif in Derlin-1 rhomboid pseudoprotease.
FEBS Lett., 590, 2016
|
|
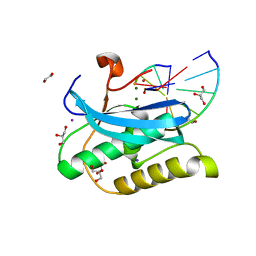
6DOK
 
 | |
9F3Q
 
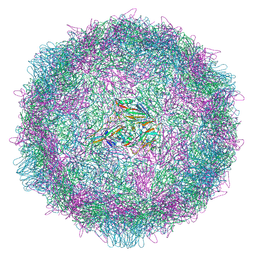 | | Poliovirus type 1 (strain Mahoney) stabilised virus-like particle (PV1 SC6b) in complex with GPP3 and GSH. | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein VP0, Capsid protein VP1, ... | | Authors: | Bahar, M.W, Nasta, V, Sherry, L, Stonehouse, N.J, Rowlands, D.J, Fry, E.E, Stuart, D.I. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
5WCZ
 
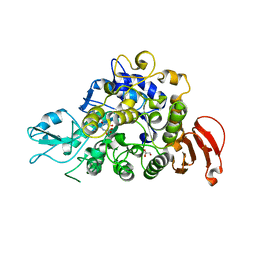 | |
