1FO3
 
 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE IN COMPLEX WITH KIFUNENSINE | | Descriptor: | ALPHA1,2-MANNOSIDASE, CALCIUM ION, KIFUNENSINE, ... | | Authors: | Vallee, F, Karaveg, K, Moremen, K.W, Herscovics, A, Howell, P.L. | | Deposit date: | 2000-08-24 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
5V82
 
 | | PIM1 kinase in complex with Cpd17 (1-(6-(4,4-difluoropiperidin-3-yl)pyridin-2-yl)-6-(6-methylpyrazin-2-yl)-1H-pyrazolo[4,3-c]pyridine) | | Descriptor: | 1-{6-[(3R)-4,4-difluoropiperidin-3-yl]pyridin-2-yl}-6-(6-methylpyrazin-2-yl)-1H-pyrazolo[4,3-c]pyridine, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Discovery of 5-Azaindazole (GNE-955) as a Potent Pan-Pim Inhibitor with Optimized Bioavailability.
J. Med. Chem., 60, 2017
|
|
1PT7
 
 | | Crystal structure of the apo-form of the yfdW gene product of E. coli | | Descriptor: | GLYCEROL, Hypothetical protein yfdW, PHOSPHATE ION | | Authors: | Gruez, A, Roig-Zamboni, V, Valencia, C, Campanacci, V, Cambillau, C. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the Escherichia coli yfdW gene product reveals a New fold of two interlaced rings identifying a wide family of CoA transferases.
J.Biol.Chem., 278, 2003
|
|
5V8Q
 
 | | Synthesis and biological evaluation of novel selective androgen receptor modulators (SARMs): Part III | | Descriptor: | 4-[(2S,3S)-2-ethyl-3-hydroxy-5-oxopyrrolidin-1-yl]-2-(trifluoromethyl)benzonitrile, Androgen receptor, GLYCEROL | | Authors: | Wilson, K.P. | | Deposit date: | 2017-03-22 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Synthesis and biological evaluation of novel selective androgen receptor modulators (SARMs) Part III: Discovery of 4-(5-oxopyrrolidine-1-yl)benzonitrile derivative 2f as a clinical candidate.
Bioorg. Med. Chem., 25, 2017
|
|
5TIL
 
 | |
1RK3
 
 | | crystal structure of the rat vitamin D receptor ligand binding domain complexed with 1,25-dihydroxyvitamin D3 and a synthetic peptide containing the NR2 box of DRIP 205 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Peroxisome proliferator-activated receptor binding protein, Vitamin D3 receptor | | Authors: | Vanhooke, J.L, M Benning, M, Bauer, C.B, Pike, J.W, DeLuca, H.F. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Structure of the Rat Vitamin D Receptor Ligand Binding Domain Complexed with 2-Carbon-Substituted Vitamin D(3) Hormone Analogues and a LXXLL-Containing Coactivator Peptide
Biochemistry, 43, 2004
|
|
5LPX
 
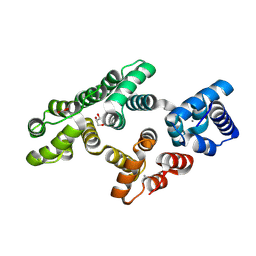 | | Crystal structure of PKC phosphorylation-mimicking mutant (S26E) Annexin A2 | | Descriptor: | Annexin A2, CALCIUM ION, GLYCEROL | | Authors: | Ecsedi, P, Gogl, G, Kiss, B, Nyitray, L. | | Deposit date: | 2016-08-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of the Equilibrium between Closed and Open Conformations of Annexin A2 by N-Terminal Phosphorylation and S100A4-Binding.
Structure, 25, 2017
|
|
5TMZ
 
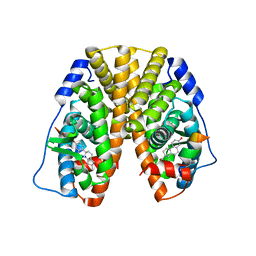 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the estradiol derivative, (8S,9S,13S,14S,17S)-16-(3-methoxybenzyl)-13-methyl-7,8,9,11,12,13,14,15,16,17-decahydro-6H-cyclopenta[a]phenanthrene-3,17-diol | | Descriptor: | (9beta,13alpha,14beta,16alpha,17alpha)-16-[(4-methoxyphenyl)methyl]estra-1,3,5(10)-triene-3,17-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
4TYG
 
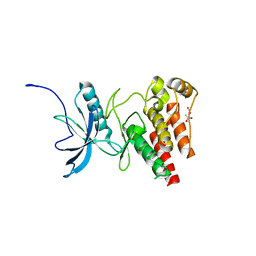 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 Kinase | | Descriptor: | ACETATE ION, Fibroblast growth factor receptor 4 | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
5TN7
 
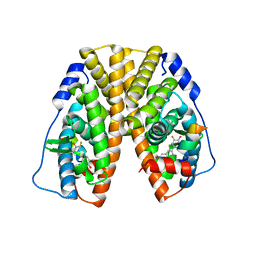 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with (E)-3'-fluoro-4'-hydroxy-3-((hydroxyiminio)methyl)-[1,1'-biphenyl]-4-olate | | Descriptor: | 3-fluoro-3'-[(E)-(hydroxyimino)methyl][1,1'-biphenyl]-4,4'-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Minutolo, F, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
1RW5
 
 | |
4UFU
 
 | | Crystal structure of human tankyrase 2 in complex with TA-12 | | Descriptor: | 8-methyl-2-[4-(trifluoromethyl)phenyl]-3H-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-19 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
5LN8
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica, in complex with galactose | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA, beta-D-galactopyranose | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LND
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-04 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
4UST
 
 | |
4US2
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 3-[(3R)-1-ethyl-2,5-dioxopyrrolidin-3-yl]benzamide, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
5LJM
 
 | | Structure of SPATA2 PUB domain | | Descriptor: | GLYCEROL, Spermatogenesis-associated protein 2 | | Authors: | Elliott, P.R, Komander, D. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
4USP
 
 | | X-ray structure of the dimeric CCL2 lectin in native form | | Descriptor: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
1MQ2
 
 | | Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*(8OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(2DA))-3', ... | | Authors: | Krahn, J.M, Beard, W.A, Miller, H, Grollman, A.P, Wilson, S.H. | | Deposit date: | 2002-09-13 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of DNA Polymerase beta with the Mutagenic DNA Lesion 8-oxodeoxyguanine Reveals
Structural Insights into its Coding Potential
Structure, 11, 2003
|
|
5LC7
 
 | |
5TBM
 
 | | Crystal structure of PT2385 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Small-Molecule Antagonist of HIF2 alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma.
Cancer Res., 76, 2016
|
|
5T4X
 
 | | CRYSTAL STRUCTURE OF PDE6D IN APO-STATE | | Descriptor: | Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Qureshi, B.M, Schmidt, A, Scheerer, P. | | Deposit date: | 2016-08-30 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mechanistic insights into the role of prenyl-binding protein PrBP/ delta in membrane dissociation of phosphodiesterase 6.
Nat Commun, 9, 2018
|
|
5LGP
 
 | | Crystal structure of mouse CARM1 in complex with ligand P1C3s | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LJ2
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 5-(5-aminopyridin-3-yl)-8-(((3R,4R)-3-((1,1-dioxidotetrahydro-2H-thiopyran-4-yl)methoxy)piperidin-4-yl)amino)-3-methyl-1,7-naphthyridin-2(1H)-one | | Descriptor: | 1,2-ETHANEDIOL, 5-(5-aminopyridin-3-yl)-8-(((3R,4R)-3-((1,1-dioxidotetrahydro-2H-thiopyran-4-yl)methoxy)piperidin-4-yl)amino)-3-methyl-1,7-naphthyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | A Chemical Probe for the ATAD2 Bromodomain.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1MFI
 
 | | CRYSTAL STRUCTURE OF MACROPHAGE MIGRATION INHIBITORY FACTOR COMPLEXED WITH (E)-2-FLUORO-P-HYDROXYCINNAMATE | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, PROTEIN (MACROPHAGE MIGRATION INHIBITORY FACTOR) | | Authors: | Taylor, A.B, Johnson Jr, W.H, Czerwinski, R.M, Whitman, C.P, Hackert, M.L. | | Deposit date: | 1998-08-12 | | Release date: | 1999-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of macrophage migration inhibitory factor complexed with (E)-2-fluoro-p-hydroxycinnamate at 1.8 A resolution: implications for enzymatic catalysis and inhibition.
Biochemistry, 38, 1999
|
|
