6JJP
 
 | | Crystal structure of Fab of a PD-1 monoclonal antibody MW11-h317 in complex with PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of MW11-h317, Programmed cell death protein 1, ... | | Authors: | Wang, M, Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation.
Commun Biol, 2, 2019
|
|
6O3O
 
 | | Structure of human DNAM-1 (CD226) bound to nectin-like protein-5 (necl-5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD226 antigen, ... | | Authors: | Deuss, F.A, Watson, G.M, Rossjohn, J, Berry, R. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the recognition of nectin-like protein-5 by the human-activating immune receptor, DNAM-1.
J.Biol.Chem., 294, 2019
|
|
6R2L
 
 | | NYBR1-A2-SLSKILDTV | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sami, M. | | Deposit date: | 2019-03-18 | | Release date: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NYBR1-A2-SLSKILDTV
To Be Published
|
|
6JO7
 
 | | Crystal structure of mouse MXRA8 | | Descriptor: | Matrix remodeling-associated protein 8 | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6JO8
 
 | | The complex structure of CHIKV envelope glycoprotein bound to human MXRA8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHIKV E1, ... | | Authors: | Song, H, Zhao, Z, Qi, J, Gao, F, Gao, F.G. | | Deposit date: | 2019-03-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Molecular Basis of Arthritogenic Alphavirus Receptor MXRA8 Binding to Chikungunya Virus Envelope Protein.
Cell, 177, 2019
|
|
6R3K
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2~{S},4~{R})-1-[[5-chloranyl-2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6OGE
 
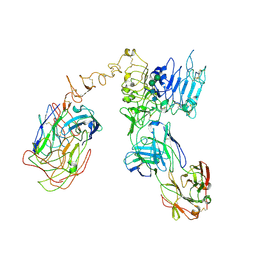 | | Cryo-EM structure of Her2 extracellular domain-Trastuzumab Fab-Pertuzumab Fab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab FAB HEAVY CHAIN, Pertuzumab FAB LIGHT CHAIN, ... | | Authors: | Hao, Y, Yu, X, Bai, Y, Huang, X. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM Structure of HER2-trastuzumab-pertuzumab complex.
Plos One, 14, 2019
|
|
6RAF
 
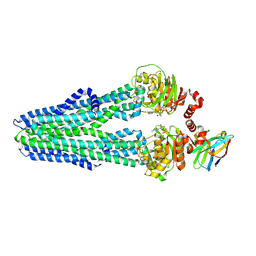 | | Heterodimeric ABC exporter TmrAB in inward-facing narrow conformation under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anti-vesicular stomatitis virus N VHH, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6OIL
 
 | | Crystal structure of human VISTA extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | Authors: | Mehta, N, Cochran, J.R, Mathews, I.I. | | Deposit date: | 2019-04-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Functional Binding Epitope of V-domain Ig Suppressor of T Cell Activation.
Cell Rep, 28, 2019
|
|
6OKQ
 
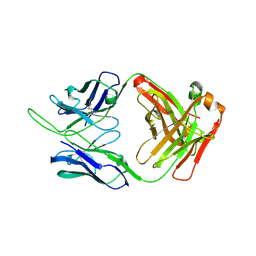 | | Crystal structure of the SF12 Fab | | Descriptor: | SF12 Fab Heavy Chain,SF12 Fab Heavy Chain, SF12 Fab Light Chain,SF12 Fab Light Chain | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2019-04-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad and Potent Neutralizing Antibodies Recognize the Silent Face of the HIV Envelope.
Immunity, 50, 2019
|
|
6OKP
 
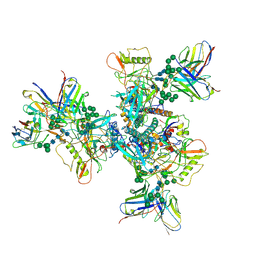 | | B41 SOSIP.664 in complex with the silent-face antibody SF12 and V3-targeting antibody 10-1074 | | Descriptor: | 10-1074 Heavy Chain,10-1074 Heavy Chain, 10-1074 Light Chain,10-1074 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2019-04-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Broad and Potent Neutralizing Antibodies Recognize the Silent Face of the HIV Envelope.
Immunity, 50, 2019
|
|
6ONB
 
 | | Crystal Structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, monoclinic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NeuRonal IgCAM-5, ... | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ON9
 
 | | Crystal Structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, tetragonal form | | Descriptor: | NeuRonal IgCAM-5, SODIUM ION, SULFATE ION, ... | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OOR
 
 | | Structure of 1B1 bound to mouse CD1d | | Descriptor: | Antibody 1B1 Heavy chain, Antibody 1B1 Light chain, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Ying, G, Zajonc, D.M. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of NKT cell inhibition using the T-cell receptor-blocking anti-CD1d antibody 1B1.
J.Biol.Chem., 294, 2019
|
|
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
6ORT
 
 | | Crystal Structure of Bos taurus Mxra8 Ectodomain | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Matrix remodeling-associated protein 8 | | Authors: | Fremont, D.H, Kim, A.S, Nelson, C.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Evolutionary Insertion in the Mxra8 Receptor-Binding Site Confers Resistance to Alphavirus Infection and Pathogenesis.
Cell Host Microbe, 27, 2020
|
|
6K0Y
 
 | | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor | | Descriptor: | 1,2-ETHANEDIOL, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Liu, J.X, Wang, G.Q. | | Deposit date: | 2019-05-08 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Study of the interactions of a novel monoclonal antibody, mAb059c, with the hPD-1 receptor.
Sci Rep, 9, 2019
|
|
6RPJ
 
 | |
6RPG
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[4-[[3-[3-[[4-[(2-acetamidoethylamino)methyl]-5-[(5-cyanopyridin-3-yl)methoxy]-2-methyl-phenoxy]methyl]-2-methyl-phenyl]-2-methyl-phenyl]methoxy]-2-[(5-cyanopyridin-3-yl)methoxy]-5-methyl-phenyl]methylamino]ethyl]ethanamide | | Authors: | Magiera-Mularz, K, Basu, S, Yang, J, Xu, B, Skalniak, L, Musielak, B, Kholodovych, V, Holak, T.A, Hu, L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, Evaluation, and Structural Studies ofC2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6RP8
 
 | |
6RQM
 
 | |
6P4B
 
 | | HyHEL10 fab variant HyHEL10-4x (heavy chain mutations L4F, Y33H, S56N, and Y58F) bound to hen egg lysozyme variant HEL2x-flex (mutations R21Q, R73E, C76S, and C94S) | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4C
 
 | | HyHEL10 Fab carrying four heavy chain mutations (HyHEL10-4x): L4F, Y33H, S56N, and Y58F | | Descriptor: | CHLORIDE ION, HyHEL10 Fab heavy chain, HyHEL10 Fab light chain | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational diversity facilitates antibody mutation trajectories and discrimination between foreign and self-antigens.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6P4A
 
 | |
6PLL
 
 | | Crystal structure of the ZIG-8 IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Zwei Ig domain protein zig-8 | | Authors: | Cheng, S, Ozkan, E. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
