5EDH
 
 | | human PDE10A, 8-ethyl-5-methyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-c]pyrimidine, 2.03A, H3, Rfree=22.7% | | Descriptor: | 8-ethyl-5-methyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-c]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
5EDI
 
 | | human PDE10A, 6-Chloro-5,8-dimethyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-2H-[1,2,4]triazol-3-yl)-ethyl]-[1,2,4]triazolo[1,5-a]pyridine, 2.20A, H3, Rfree=23.5% | | Descriptor: | 6-chloranyl-5,8-dimethyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-a]pyridine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
4AXF
 
 | | InsP5 2-K in complex with Ins(3,4,5,6)P4 plus AMPPNP | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, Myo inositol 3,4,5,6 tetrakisphosphate, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4AXD
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with AMPPNP | | Descriptor: | CITRIC ACID, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
7JZ2
 
 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
7JZ6
 
 | |
6TNH
 
 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with AMPPcP, dGMP, Asp, Magnesium at 2.21 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ASPARTIC ACID, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
6W18
 
 | | Structure of S. pombe Arp2/3 complex in inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1, ... | | Authors: | Shaaban, M, Nolen, B.J, Chowdhury, S. | | Deposit date: | 2020-03-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JZH
 
 | |
7JZ3
 
 | | Osmoporin OmpC from E.coli K12 | | Descriptor: | Outer membrane protein C | | Authors: | Lyu, M, Su, C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
6U2H
 
 | | BRAF dimer bound to 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1W49
 
 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP and Mg | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W48
 
 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
5LLF
 
 | |
6O7I
 
 | |
1W4A
 
 | | P4 protein from PHI12 in complex with AMPcPP and Mn | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
8UYI
 
 | | Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase Pink1, ... | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
8UYF
 
 | | Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | Serine/threonine-protein kinase Pink1, mitochondrial | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
4BLV
 
 | | Crystal structure of Escherichia coli 23S rRNA (A2030-N6)- methyltransferase RlmJ in complex with S-adenosylmethionine (AdoMet) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Punekar, A.S, Liljeruhm, J, Shepherd, T.R, Forster, A.C, Selmer, M. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights Into the Molecular Mechanism of Rrna M6A Methyltransferase Rlmj.
Nucleic Acids Res., 41, 2013
|
|
4CO4
 
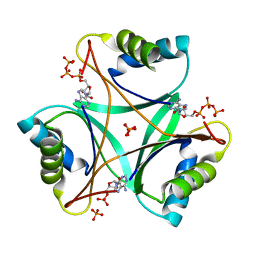 | | Structure of PII signaling protein GlnZ from Azospirillum brasilense in complex with adenosine triphosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Truan, D, Li, X.-D, Winkler, F.K. | | Deposit date: | 2014-01-25 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Thermodynamics of Effector Molecule Binding to the Nitrogen Signal Transduction Pii Protein Glnz from Azospirillum Brasilense.
J.Mol.Biol., 426, 2014
|
|
3UI7
 
 | | Discovery of orally active pyrazoloquinoline as a potent PDE10 inhibitor for the management of schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(morpholin-4-ylmethyl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, S, Smotryski, J, Mcelroy, W, Ho, G, Tulshian, D, Greenlee, W.J, Hodgson, R, Xiao, L, Hruza, A. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of orally active pyrazoloquinolines as potent PDE10 inhibitors for the management of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4D79
 
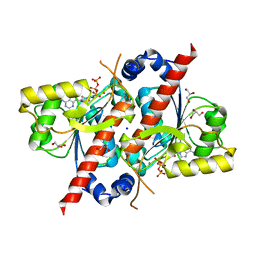 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with ATP at 1.768 Angstroem resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
6SIC
 
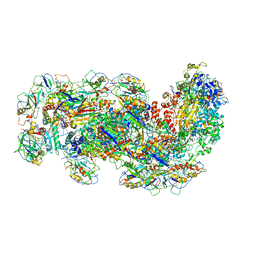 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | Descriptor: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-08-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
5N23
 
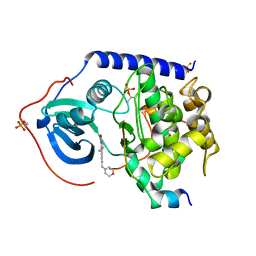 | |
4AZS
 
 | | High resolution (2.2 A) crystal structure of WbdD. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, METHYLTRANSFERASE WBDD, ... | | Authors: | Hagelueken, G, Huang, H, Naismith, J.H. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Wbdd; a Bifunctional Kinase and Methyltransferase that Regulates the Chain Length of the O Antigen in Escherichia Coli O9A.
Mol.Microbiol., 86, 2012
|
|
