6X9H
 
 | | Molecular mechanism and structural basis of small-molecule modulation of acid-sensing ion channel 1 (ASIC1) | | Descriptor: | 2-[4-(3,4-dimethoxyphenoxy)phenyl]-1H-benzimidazole-6-carboximidamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, ... | | Authors: | Liu, Y, Ma, J, DesJarlais, R.L, Hagan, R, Rech, J, Lin, D, Liu, C, Miller, R, Schoellerman, J, Luo, J, Letavic, M, Grasberger, B, Maher, M. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular mechanism and structural basis of small-molecule modulation of the gating of acid-sensing ion channel 1.
Commun Biol, 4, 2021
|
|
5NRD
 
 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-2 | | Descriptor: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5NOL
 
 | | Ca2+-induced Movement of Tropomyosin on Native Cardiac Thin Filaments - "Closed" state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cardiac muscle alpha actin 1, MAGNESIUM ION, ... | | Authors: | Risi, C, Eisner, J, Belknap, B, Heeley, D.H, White, H.D, Schroeder, G.F, Galkin, V.E. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Ca(2+)-induced movement of tropomyosin on native cardiac thin filaments revealed by cryoelectron microscopy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7Q8B
 
 | | Leishmania major actin filament in ADP-Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION, ... | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
7QHM
 
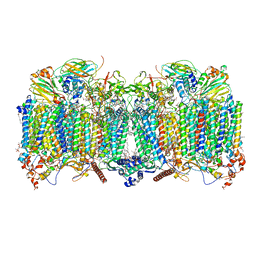 | | Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (stigmatellin and azide bound) | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, AZIDE ION, ... | | Authors: | Kao, W.-C, Hunte, C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for safe and efficient energy conversion in a respiratory supercomplex.
Nat Commun, 13, 2022
|
|
7QHO
 
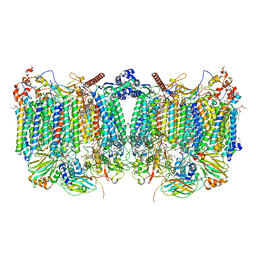 | | Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (as isolated) | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Kao, W.-C, Hunte, C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for safe and efficient energy conversion in a respiratory supercomplex
Nature Communications, 13, 2022
|
|
5DCG
 
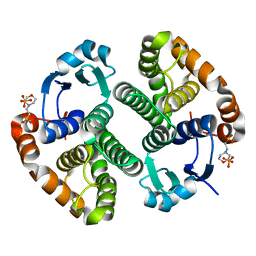 | |
7Q8C
 
 | | Leishmania major actin filament in ADP-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
6XD9
 
 | | Carbonmonoxy hemoglobin in complex with the antisickling agent 2-hydroxy-6-((6-(hydroxymethyl)pyridin-2-yl)methoxy)benzaldehyde (VZHE039) | | Descriptor: | 3-{[6-(hydroxymethyl)pyridin-2-yl]methoxy}-2-methylphenol, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Pagare, P.P, Safo, M.K, Musayev, F.N. | | Deposit date: | 2020-06-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | VZHE-039, a novel antisickling agent that prevents erythrocyte sickling under both hypoxic and anoxic conditions.
Sci Rep, 10, 2020
|
|
7Q8S
 
 | | Leishmania major ADP-actin filament decorated with Leishmania major cofilin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADF/Cofilin, Actin, ... | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
6XDT
 
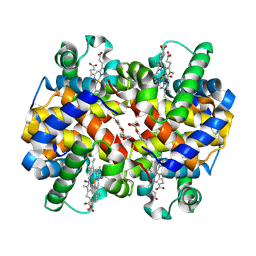 | | Carbonmonoxy hemoglobin in complex with the antisickling agent methyl 5-((2-formyl-4-methoxyphenoxy)methyl)picolinate | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Pagare, P.P, Safo, M.K, Musayev, F.N. | | Deposit date: | 2020-06-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploration of Structure-Activity Relationship of Aromatic Aldehydes Bearing Pyridinylmethoxy-Methyl Esters as Novel Antisickling Agents.
J.Med.Chem., 63, 2020
|
|
4RPP
 
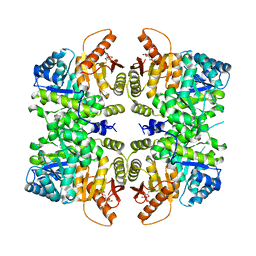 | | crystal structure of PKM2-K422R mutant bound with FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Pyruvate kinase PKM | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
5H7H
 
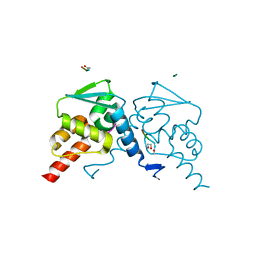 | | Crystal structure of the BCL6 BTB domain in complex with F1324(10-13) | | Descriptor: | 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, F1324 peptide residues 10-13 | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2016-11-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of high-affinity BCL6-binding peptide and its structure-activity relationship.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
3N2R
 
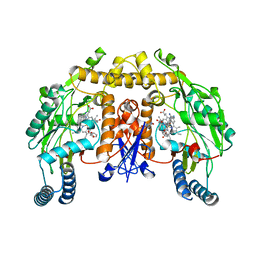 | | Structure of neuronal nitric oxide synthase heme domain in complex with 6-(((3R,4R/3S,4S)-4-(3-Phenoxyphenoxy)pyrrolidin-3-yl)methyl)pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-(3-phenoxyphenoxy)pyrrolidin-3-yl]methyl}pyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2010-05-18 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of lipophilic-tailed monocationic inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 18, 2010
|
|
6XDK
 
 | |
3N5P
 
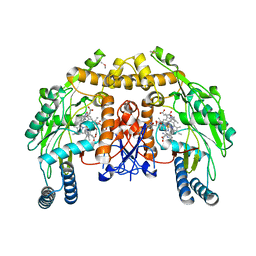 | | Structure of endothelial nitric oxide synthase heme domain complexed with 4-(2-(6-(2-(6-amino-4-methylpyridin-2-yl)ethyl)pyridin-2-yl)ethyl)-6-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{6-[2-(2-amino-6-methylpyridin-4-yl)ethyl]pyridin-2-yl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
5NR9
 
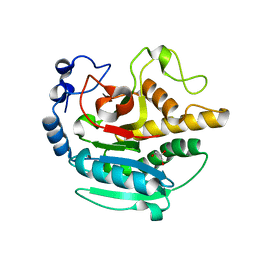 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (alpha-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - Unliganded alpha-3GalT | | Descriptor: | GLYCEROL, N-acetyllactosaminide alpha-1,3-galactosyltransferase | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3N62
 
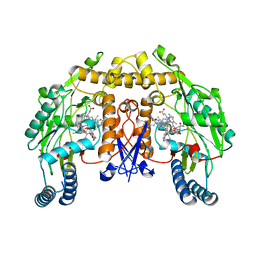 | | Structure of neuronal nitric oxide synthase D597N mutant heme domain in complex with 6,6'-(2,2'-(pyridine-3,5-diyl)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-(pyridine-3,5-diyldiethane-2,1-diyl)bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Li, H, Delker, S.L, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
7QRG
 
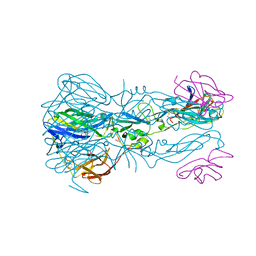 | |
5DFJ
 
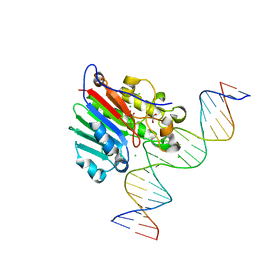 | | Human APE1 E96Q/D210N mismatch substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5I73
 
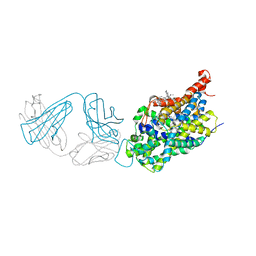 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central and allosteric sites | | Descriptor: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
6XGF
 
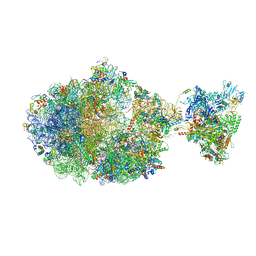 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 30 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-17 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
5NRA
 
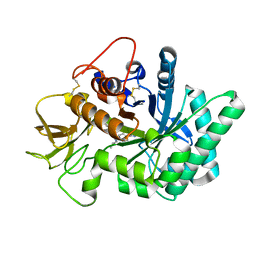 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7g | | Descriptor: | 1-(5-azanyl-4~{H}-1,2,4-triazol-3-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-(2-methylpropyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.267 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
5NRF
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7i | | Descriptor: | 1-(3-azanyl-1~{H}-1,2,4-triazol-5-yl)-~{N}-[2-(4-chlorophenyl)ethyl]-~{N}-(phenylmethyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
7QSM
 
 | | Bovine complex I in lipid nanodisc, Deactive-ligand (composite) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Chung, I, Bridges, H.R, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-25 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
