1AUZ
 
 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BHI
 
 | | STRUCTURE OF TRANSACTIVATION DOMAIN OF CRE-BP1/ATF-2, NMR, 20 STRUCTURES | | Descriptor: | CRE-BP1 | | Authors: | Nagadoi, A, Nakazawa, K, Uda, H, Maekawa, T, Ishii, S, Nishimura, Y. | | Deposit date: | 1998-06-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain.
J.Mol.Biol., 287, 1999
|
|
8BZ9
 
 | | single soak stabilizer for ERa - 14-3-3 interaction (AZ354) | | Descriptor: | 14-3-3 protein sigma, 4-chloranyl-7-propan-2-yloxy-1-benzothiophene-2-carboximidamide, ERalpha peptide, ... | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BYG
 
 | | fragment-linked stabilizer for ERa - 14-3-3 interaction (1047648) | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, ~{N}-[2-[(2-carbamimidoyl-1-benzothiophen-4-yl)-methyl-amino]ethyl]-2-(4-chloranylphenoxy)-~{N},2-dimethyl-propanamide | | Authors: | Visser, E.J, Sijbesma, E, Ottmann, C. | | Deposit date: | 2022-12-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From Tethered to Freestanding Stabilizers of 14-3-3 Protein-Protein Interactions through Fragment Linking.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7NR4
 
 | | X-RAY STRUCTURE OF PRMT6 IN COMPLEX WITH indazole type inhibitor | | Descriptor: | (2~{S})-2-azanyl-~{N}-[3-[3-(dimethylsulfamoyl)phenyl]-2~{H}-indazol-5-yl]propanamide, Protein arginine N-methyltransferase 6, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Steuber, H. | | Deposit date: | 2021-03-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational Design and Synthesis of Selective PRMT4 Inhibitors: A New Chemotype for Development of Cancer Therapeutics*.
Chemmedchem, 16, 2021
|
|
8CAF
 
 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | Descriptor: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
1BI8
 
 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURES CDK6-P19INK4D INHIBITOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, CYCLIN-DEPENDENT KINASE INHIBITOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
8C2E
 
 | |
8C2F
 
 | |
7NVQ
 
 | | Aerosol-soaked human cdk2 crystals with Staurosporine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, STAUROSPORINE | | Authors: | Ross, B, Huber, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Aerosol-based ligand soaking of reservoir-free protein crystals.
J.Appl.Crystallogr., 54, 2021
|
|
7O07
 
 | |
8BZF
 
 | | FC-J acetonide stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, Fusicoccin J-acetonide, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
8BYX
 
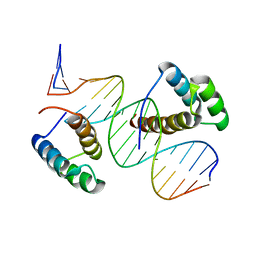 | |
8BZC
 
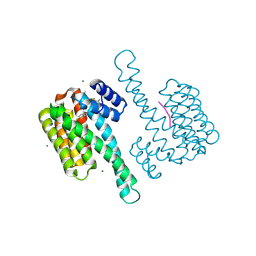 | |
8BZM
 
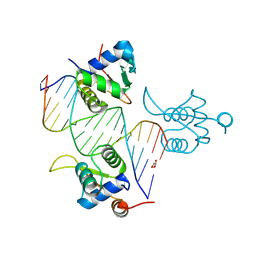 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | FOXK1-ELF1_heterodimer bound to DNA
To Be Published
|
|
7O7K
 
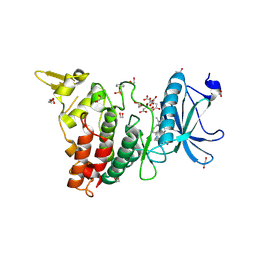 | | Crystal structure of the human DYRK1A kinase domain bound to abemaciclib | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
7O7I
 
 | |
8C28
 
 | | 14-3-3 in complex with PyrinpS208pS242 | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyrin | | Authors: | Lau, R, Ottmann, C, Hann, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 14-3-3/Pyrin complex
To Be Published
|
|
7O7J
 
 | | Crystal structure of the human HIPK3 kinase domain bound to abemaciclib | | Descriptor: | Homeodomain-interacting protein kinase 3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
8C30
 
 | | Crystal structure of 14-3-3 in complex with PyrinpS242 and a protein/peptide interface fragment | | Descriptor: | 14-3-3 protein sigma, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, ... | | Authors: | Lau, R, Ottmann, C. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 14-3-3/Pyrin complex
To Be Published
|
|
8C2Y
 
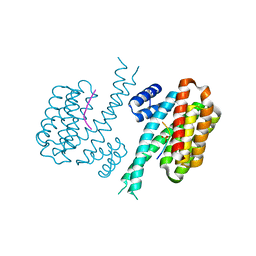 | |
8COG
 
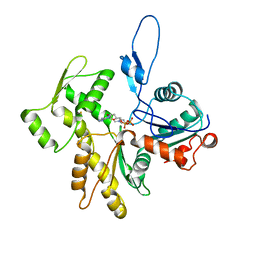 | | Human arginylated beta-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Pinto, C.S, Bakker, S.E, Suchenko, A, Hussain, H, Hatano, T, Sampath, K, Chinthalapudi, K, Mishima, M, Balasubramanian, M. | | Deposit date: | 2023-02-28 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.499 Å) | | Cite: | Structure and physiological investigation of human arginylated beta-actin
To Be Published
|
|
7NUE
 
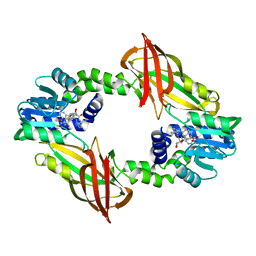 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML736 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[5-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]pentylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
7NUD
 
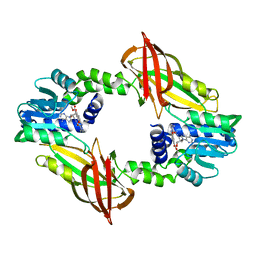 | | Crystal structure of mouse PRMT6 in complex with inhibitor EML734 | | Descriptor: | Protein arginine N-methyltransferase 6, methyl 6-[3-[[~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]carbamimidoyl]amino]propylcarbamoylamino]-4-oxidanyl-naphthalene-2-carboxylate | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.
J.Med.Chem., 65, 2022
|
|
1BUZ
 
 | | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SPOIIAA | | Authors: | Kovacs, H, Comfort, D, Lord, M, Campbell, I.D, Yudkin, M.D. | | Deposit date: | 1997-09-08 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
