6R77
 
 | |
4UJ1
 
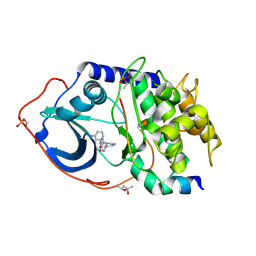 | | Protein Kinase A in complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(3S,4R)-4-(3-chlorophenyl)carbonylpyrrolidin-3-yl]-3H-quinazolin-4-one, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of Pka and a Pkb Mimic.
Chemistry, 22, 2016
|
|
6QYD
 
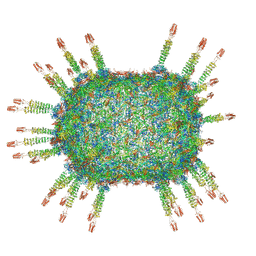 | | Cryo-EM structure of the head in mature bacteriophage phi29 | | Descriptor: | Capsid fiber protein, Major capsid protein | | Authors: | Xu, J.W, Wang, D.H, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
4ULV
 
 | | Cytochrome c prime from Shewanella frigidimarina | | Descriptor: | CYTOCHROME C, CLASS II, GLYCEROL, ... | | Authors: | Manole, A.A, Kekilli, D, Dobbin, P.S, Hough, M.A. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Conformational Control of the Binding of Diatomic Gases to Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
7RYU
 
 | |
4UMD
 
 | |
4UNT
 
 | | Induced monomer of the Mcg variable domain | | Descriptor: | IG LAMBDA CHAIN V-II REGION MGC, SULFATE ION | | Authors: | Brumshtein, B, Esswein, S.R, Landau, M, Ryan, C.M, Whitelegge, J.P, Phillips, M.L, Cascio, D, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2014-05-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Formation of Amyloid Fibers by Monomeric Light-Chain Variable Domains.
J.Biol.Chem., 289, 2014
|
|
6R7R
 
 | | Crystal structure of the glutamate transporter homologue GltTk in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, DECYL-BETA-D-MALTOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Arkhipova, V, Guskov, A, Slotboom, D.J. | | Deposit date: | 2019-03-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and transport of D-aspartate by the glutamate transporter homolog Glt Tk .
Elife, 8, 2019
|
|
4UNS
 
 | | Mtb TMK in complex with compound 40 | | Descriptor: | N-[4-(3-CYANO-7-ETHYL-5-METHYL-2-OXO-1H-1,6-NAPHTHYRIDIN-4-YL)PHENYL]METHANESULFONAMIDE, SODIUM ION, THYMIDYLATE KINASE | | Authors: | Read, J.A, Hussein, S, Gingell, H, Tucker, J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure Guided Lead Generation for M. Tuberculosis Thymidylate Kinase (Mtb Tmk): Discovery of 3-Cyanopyridone and 1,6-Naphthyridin-2-One as Potent Inhibitors.
J.Med.Chem., 58, 2015
|
|
4P5R
 
 | | Structure of oxidized W45Y mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, SODIUM ION | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The sole tryptophan of amicyanin enhances its thermal stability but does not influence the electronic properties of the type 1 copper site.
Arch.Biochem.Biophys., 550-551, 2014
|
|
4UP9
 
 | |
4UHO
 
 | | Characterization of a Novel Transaminase from Pseudomonas sp. Strain AAC | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Wilding, M, Peat, T.S, Newman, J, Scott, C. | | Deposit date: | 2015-03-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A Beta-Alanine Catabolism Pathway Containing a Highly Promiscuous Omega-Transaminase in the 12-Aminododecanate-Degrading Pseudomonas Sp. Strain Aac.
Appl.Environ.Microbiol., 82, 2016
|
|
7SEN
 
 | |
4UMB
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UGW
 
 | | Structure of Y357F Bacillus subtilis Nitric Oxide Synthase in complex with 6-(5-((3R,4R)-4-((6-azanyl-4-methyl-pyridin-2-yl)methyl) pyrrolidin-3-yl)oxypentyl)-4-methyl-pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[5-({(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}oxy)pentyl]-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
7DBI
 
 | | Crystal structure of the peroxisomal acyl-CoA hydrolase MpaH | | Descriptor: | acyl-CoA hydrolase MpaH' | | Authors: | Li, S.Y, You, C. | | Deposit date: | 2020-10-20 | | Release date: | 2021-04-28 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for substrate specificity of the peroxisomal acyl-CoA hydrolase MpaH' involved in mycophenolic acid biosynthesis.
Febs J., 288, 2021
|
|
4UMI
 
 | |
2RG3
 
 | | Covalent complex structure of elastase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Leukocyte elastase | | Authors: | Huang, W, Yamamoto, Y. | | Deposit date: | 2007-10-02 | | Release date: | 2008-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshot of the mechanism of inactivation of human neutrophil elastase by 1,2,5-thiadiazolidin-3-one 1,1-dioxide derivatives.
J.Med.Chem., 51, 2008
|
|
6R8J
 
 | |
4UM7
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (kdsC) from Moraxella catarrhalis in complex with Magnesium ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6LEO
 
 | | Crystal structure of thiosulfate transporter YeeE from Spirochaeta thermophila | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Sulf_transp domain-containing protein, THIOSULFATE | | Authors: | Tanaka, Y, Tsukazaki, T, Yoshikaie, K, Takeuchi, A, Uchino, S, Sugano, Y. | | Deposit date: | 2019-11-26 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a YeeE/YedE family protein engaged in thiosulfate uptake.
Sci Adv, 6, 2020
|
|
4P6T
 
 | | Crystal Structure of tyrosinase from Bacillus megaterium with p-tyrosol in the active site | | Descriptor: | 4-(2-hydroxyethyl)phenol, COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2014-03-25 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins.
Nat Commun, 5, 2014
|
|
6R8L
 
 | | Human Cyclophilin D in complex with 1-((1S,9S,10S)-10-Hydroxy-12-oxa-8-aza-tricyclo[7.3.1.02,7]trideca-2,4,6-trien-4-ylmethyl)-3- {2-[(R)-2-(2-methylsulfanyl-phenyl)-pyrrolidin-1-yl]-2-oxo-ethyl}-urea | | Descriptor: | 1-[2-[(2~{R})-2-(2-methylsulfanylphenyl)pyrrolidin-1-yl]-2-oxidanylidene-ethyl]-3-[[(1~{S},9~{S},10~{S})-10-oxidanyl-12-oxa-8-azatricyclo[7.3.1.0^{2,7}]trideca-2(7),3,5-trien-4-yl]methyl]urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Graedler, U. | | Deposit date: | 2019-04-02 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5HRU
 
 | |
4UMA
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3 deoxy D arabino heptulosonate 7 phosphate synthase the importance of accommodating the active site water | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
