4L76
 
 | | Ca2+-bound E212Q mutant MthK RCK domain | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel MthK, SODIUM ION | | Authors: | Smith, F.J, Rothberg, B.S. | | Deposit date: | 2013-06-13 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | Structural basis of allosteric interactions among Ca(2+)-binding sites in a K(+) channel RCK domain.
Nat Commun, 4, 2013
|
|
3M70
 
 | |
4JWR
 
 | |
4L9Q
 
 | | X-ray study of human serum albumin complexed with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4JO0
 
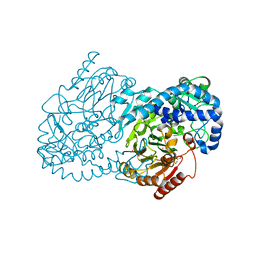 | | Crystal Structure of CmlA, a diiron beta-hydroxylase from Streptomyces venezuelae | | Descriptor: | ACETATE ION, CmlA, FE (III) ION, ... | | Authors: | Knoot, C.J, Makris, T.M, Wilmot, C.M, Lipscomb, J.D. | | Deposit date: | 2013-03-16 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of a Dinuclear Iron Cluster-Containing beta-Hydroxylase Active in Antibiotic Biosynthesis.
Biochemistry, 52, 2013
|
|
2NO2
 
 | |
4LAO
 
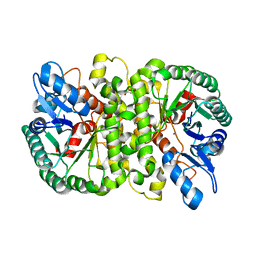 | | Crystal structure of Cordyceps militaris IDCase H195A mutant (Zn) | | Descriptor: | Cordyceps militaris IDCase, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
2IIC
 
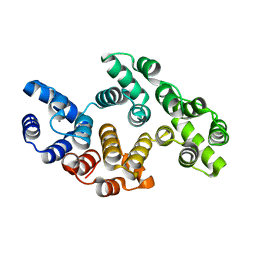 | |
2NQO
 
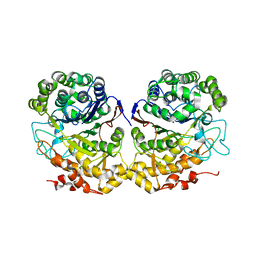 | | Crystal Structure of Helicobacter pylori gamma-Glutamyltranspeptidase | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Boanca, G, Sand, A, Okada, T, Suzuki, H, Kumagai, H, Fukuyama, K, Barycki, J.J. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoprocessing of Helicobacter pylori gamma-glutamyltranspeptidase leads to the formation of a threonine-threonine catalytic dyad.
J.Biol.Chem., 282, 2007
|
|
5MZ6
 
 | | Cryo-EM structure of a Separase-Securin complex from Caenorhabditis elegans at 3.8 A resolution | | Descriptor: | Interactor of FizzY protein, SEParase | | Authors: | Boland, A, Martin, T.G, Zhang, Z, Yang, J, Bai, X.C, Chang, L, Scheres, S.H.W, Barford, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-08 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3G0Q
 
 | | Crystal Structure of MutY bound to its inhibitor DNA | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*AP*GP*TP*CP*TP*T)-3', A/G-specific adenine glycosylase, ... | | Authors: | Lee, S, Verdine, G.L. | | Deposit date: | 2009-01-28 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3MDI
 
 | |
3FXM
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Citrate at 10 mM of Mn2+ | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3MRO
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with Melan-A MART1 decapeptide variant | | Descriptor: | 10-meric peptide from Melanoma antigen recognized by T-cells 1, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Reiser, J.-B, Le Gorrec, M, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
3MG4
 
 | | Structure of yeast 20S proteasome with Compound 1 | | Descriptor: | (2S)-2-amino-N-[(1S)-1-({(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}carbamoyl)-3-phenylpropyl]-4-phenylbutanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
3G2Z
 
 | |
4L78
 
 | | Xenon Trapping and Statistical Coupling Analysis Uncover Regions Important for Structure and Function of Multidomain Protein StPurL | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tanwar, A.S, Goyal, V.D, Choudhary, D, Panjikar, S, Anand, R. | | Deposit date: | 2013-06-13 | | Release date: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Importance of hydrophobic cavities in allosteric regulation of formylglycinamide synthetase: insight from xenon trapping and statistical coupling analysis
Plos One, 8, 2013
|
|
4LF8
 
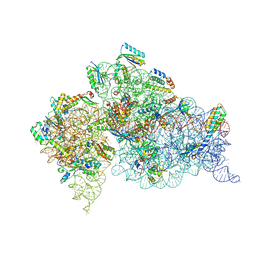 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, PAROMOMYCIN, ... | | Authors: | Demirci, H, Belardinelli, R, Carr, J, Murphy IV, F, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-06-26 | | Release date: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (3.1484 Å) | | Cite: | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus
to be published, 2013
|
|
3FYN
 
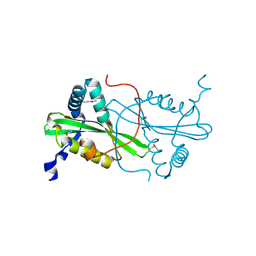 | | Crystal structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3 | | Descriptor: | ACETATE ION, Integron gene cassette protein HFX_CASS3, MAGNESIUM ION | | Authors: | Sureshan, V, Deshpande, C.N, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Osipiuk, J, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure from the mobile metagenome of Cole Harbour Salt Marsh: Integron Cassette Protein HFX_CASS3
To be Published
|
|
4J9U
 
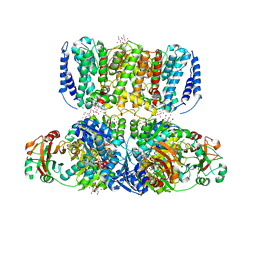 | | Crystal Structure of the TrkH/TrkA potassium transport complex | | Descriptor: | HEXATANTALUM DODECABROMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-02-17 | | Release date: | 2013-04-03 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Gating of the TrkH ion channel by its associated RCK protein TrkA.
Nature, 496, 2013
|
|
4JE4
 
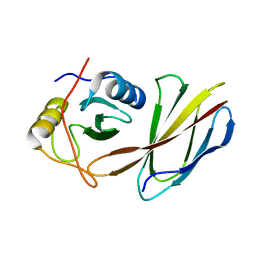 | | Crystal Structure of Monobody NSa1/SHP2 N-SH2 Domain Complex | | Descriptor: | Monobody NSa1, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sha, F, Koide, S. | | Deposit date: | 2013-02-26 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JEO
 
 | |
3GI6
 
 | | Crystal structure of protease inhibitor, AD78 in complex with wild type HIV-1 protease | | Descriptor: | (5S)-N-[(1S,2R)-2-Hydroxy-3-[[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino]-1-(phenylmethyl)propyl]-2-oxo-3-[3-(trif luoromethyl)phenyl]-5-oxazolidinecarboxamide, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
4LJ3
 
 | | Crystal structure of the EAL domain of c-di-GMP specific phosphodiesterase YahA in complex with substrate c-di-GMP and Ca++ | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Cyclic di-GMP phosphodiesterase YahA, ... | | Authors: | Sundriyal, A, Schirmer, T. | | Deposit date: | 2013-07-04 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inherent Regulation of EAL Domain-catalyzed Hydrolysis of Second Messenger Cyclic di-GMP.
J.Biol.Chem., 289, 2014
|
|
3MLU
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a ZAM18 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
