6QK4
 
 | | Lytic transglycosylase, LtgG, of Burkholderia pseudomallei. | | Descriptor: | Membrane-bound lytic murein transglycosylase A | | Authors: | Jenkins, C.H, Wallis, R, Allcock, N, Barnes, K.B, Richards, M.I, Auty, J.M, Galyov, E.E, Harding, S.V, Mukamolova, G.V. | | Deposit date: | 2019-01-28 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The lytic transglycosylase, LtgG, controls cell morphology and virulence in Burkholderia pseudomallei.
Sci Rep, 9, 2019
|
|
6QKG
 
 | | 2-Naphthoyl-CoA Reductase(NCR) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Kayastha, K, Ermler, U. | | Deposit date: | 2019-01-29 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low potential enzymatic hydride transfer via highly cooperative and inversely functionalized flavin cofactors.
Nat Commun, 10, 2019
|
|
6QKY
 
 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6QDW
 
 | | Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Hodirnau, V.V, Kudlinzki, D, Mao, J, Glaubitz, C, Frangakis, A, Schwalbe, H. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cysteine oxidation and disulfide formation in the ribosomal exit tunnel.
Nat Commun, 11, 2020
|
|
6QGM
 
 | | VirX1 apo structure | | Descriptor: | VirX1 | | Authors: | Gkotsi, D.S, Ludewig, H, Sharma, S.V, Unsworth, W.P, Taylor, R.J.K, McLachlan, M.M.W, Shanahan, S, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A marine viral halogenase that iodinates diverse substrates.
Nat.Chem., 11, 2019
|
|
6QDE
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000877 | | Descriptor: | 3-[[6-~{tert}-butyl-2-[3-(dimethylamino)propyl-methyl-amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QDU
 
 | | Crystal structure of 14-3-3sigma in complex with a RapGef2 pT740 phosphopeptide inhibited by semi-synthetic fusicoccane FC-NCPC | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, FC-NCPC, ... | | Authors: | Andrei, S.A, Kaplan, A, Fournier, A.E, Ottman, C. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Polypharmacological Perturbation of the 14-3-3 Adaptor Protein Interactome Stimulates Neurite Outgrowth.
Cell Chem Biol, 27, 2020
|
|
6QE3
 
 | | Re-refinement of 6ESR human IBA57 at 1.75 A resolution | | Descriptor: | Putative transferase CAF17, mitochondrial | | Authors: | Calderone, V, Ciofi-Baffoni, S, Gourdoupis, S, Banci, L. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In-house high-energy-remote SAD phasing using the magic triangle: how to tackle the P1 low symmetry using multiple orientations of the same crystal of human IBA57 to increase the multiplicity.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QHC
 
 | | Crystal Structure of Human Kallikrein 6 in complex with GSK358180B | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Kallikrein-6, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6QHM
 
 | | 14-3-3 sigma with RelA/p65 binding site pS281 | | Descriptor: | 14-3-3 protein sigma, LEU-SEP-GLU | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-01-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
6QHO
 
 | |
6QHP
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 2256 ms covalent intermediate 1 | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
6QEF
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR (S)-3-Hydroxy-2-oxo-1-phenyl-pyrrolidine-3-carboxylic acid 3-chloro-5-fluoro-benzylamide | | Descriptor: | (3~{S})-~{N}-[(3-chloranyl-5-fluoranyl-phenyl)methyl]-3-oxidanyl-2-oxidanylidene-1-phenyl-pyrrolidine-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6QEQ
 
 | | PcfF from Enterococcus faecalis pCF10 | | Descriptor: | PcfF | | Authors: | Rehman, S, Berntsson, R.P.A. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-01 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enterococcal PcfF Is a Ribbon-Helix-Helix Protein That Recruits the Relaxase PcfG Through Binding and Bending of the oriT Sequence.
Front Microbiol, 10, 2019
|
|
6QF5
 
 | | X-Ray structure of human Aquaporin 2 crystallized on a silicon chip | | Descriptor: | Aquaporin-2, CADMIUM ION | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QFL
 
 | |
6QFU
 
 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 7) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-oxidanyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1(6),2,4-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
6QFX
 
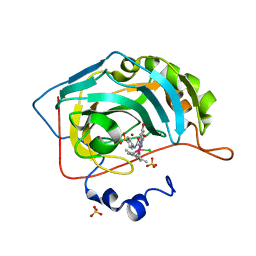 | | Human carbonic anhydrase II with bound IrCp* complex (cofactor 10) to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-oxidanyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Chemical Optimization of Whole-Cell Transfer Hydrogenation Using Carbonic Anhydrase as Host Protein.
Acs Catalysis, 9, 2019
|
|
6QG5
 
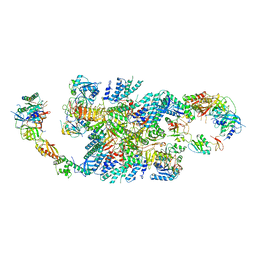 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model C) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QGR
 
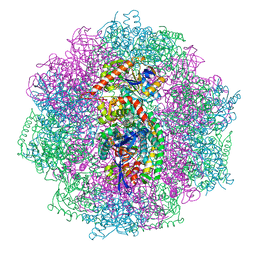 | | The F420-reducing [NiFe] hydrogenase complex from Methanosarcina barkeri at the Nia-S state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, Coenzyme F420 hydrogenase subunit alpha, ... | | Authors: | Ilina, Y, Lorent, C, Katz, S, Jeoung, J.H, Shima, S, Horch, M, Zebger, I, Dobbek, H. | | Deposit date: | 2019-01-12 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | X-ray Crystallography and Vibrational Spectroscopy Reveal the Key Determinants of Biocatalytic Dihydrogen Cycling by [NiFe] Hydrogenases.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QH7
 
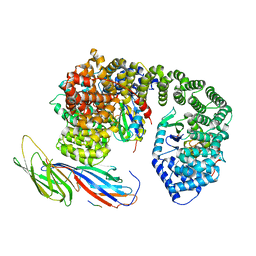 | | AP2 clathrin adaptor mu2T156-phosphorylated core with two cargo peptides in open+ conformation | | Descriptor: | ADAPTOR-RELATED PROTEIN COMPLEX 2, MU 2 SUBUNIT, C-TERMINAL DOMAIN, ... | | Authors: | Wrobel, A.G, Owen, D.J, McCoy, A.J, Evans, P.R. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6QHX
 
 | | Time resolved structural analysis of the full turnover of an enzyme - 6156 ms | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Pai, E.F, Miller, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Time-resolved crystallography reveals allosteric communication aligned with molecular breathing.
Science, 365, 2019
|
|
7TQV
 
 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA | | Descriptor: | RNA (33-MER), Uridylate-specific endoribonuclease | | Authors: | Frazier, M.N, Krahn, J.M, Butay, K.J, Dillard, L.B, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Nucleic Acids Res., 50, 2022
|
|
7TK5
 
 | |
7TKE
 
 | |
