4CRH
 
 | | Crystal structure of the BTB-T1 domain of human SHKBP1 | | Descriptor: | SH3KBP1-BINDING PROTEIN 1 | | Authors: | Pinkas, D.M, Solcan, N, Krojer, T, Goubin, S, Williams, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
4CO8
 
 | | Structure of the DNA binding ETS domain of human ETV4 | | Descriptor: | 1,2-ETHANEDIOL, ETS TRANSLOCATION VARIANT 4 | | Authors: | Newman, J.A, Cooper, C.D.O, Shrestha, L, Burgess-Brown, N, Kopec, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2014-01-27 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
3EIE
 
 | | Crystal Structure of S.cerevisiae Vps4 in the SO4-bound state | | Descriptor: | SULFATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
6WQE
 
 | | Solution Structure of the IWP-051-bound H-NOX from Shewanella woodyi in the Fe(II)CO ligation state | | Descriptor: | 5-fluoro-2-{1-[(2-fluorophenyl)methyl]-5-(1,2-oxazol-3-yl)-1H-pyrazol-3-yl}pyrimidin-4-ol, CARBON MONOXIDE, Heme NO binding domain protein, ... | | Authors: | Chen, C.Y, Lee, W, Montfort, W.R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the Shewanella woodyi H-NOX protein in the presence and absence of soluble guanylyl cyclase stimulator IWP-051.
Protein Sci., 30, 2021
|
|
1U36
 
 | | Crystal structure of WLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U94
 
 | |
4E7P
 
 | | Crystal structure of receiver domain of putative NarL family response regulator spr1814 from Streptococcus pneumoniae in the presence of the phosphoryl analog beryllofluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator | | Authors: | Park, A.K, Moon, J.H, Lee, K.S, Chi, Y.M. | | Deposit date: | 2012-03-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Crystal structure of receiver domain of putative NarL family response regulator spr1814 from Streptococcus pneumoniae in the absence and presence of the phosphoryl analog beryllofluoride.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
1U3Y
 
 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U41
 
 | | Crystal structure of YLGV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
2X7V
 
 | | Crystal structure of Thermotoga maritima endonuclease IV in the presence of zinc | | Descriptor: | PROBABLE ENDONUCLEASE 4, ZINC ION | | Authors: | Tomanicek, S.J, Hughes, R.C, Ng, J.D, Coates, L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Endonuclease Iv Homologue from Thermotoga Maritima in the Presence of Active-Site Divalent Metal Ions
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1U99
 
 | |
3FPL
 
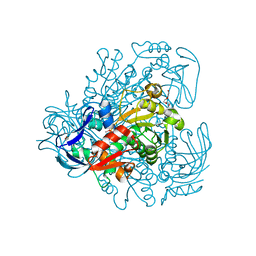 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of C. beijerinckii ADH by T. brockii ADH | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Felix, F, Goihberg, E, Shimon, L, Burstein, Y. | | Deposit date: | 2009-01-05 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
1UEL
 
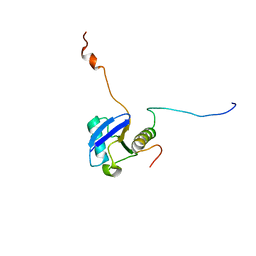 | | Solution structure of ubiquitin-like domain of hHR23B complexed with ubiquitin-interacting motif of proteasome subunit S5a | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, UV excision repair protein RAD23 homolog B | | Authors: | Fujiwara, K, Tenno, T, Jee, J.G, Sugasawa, K, Ohki, I, Kojima, C, Tochio, H, Hiroaki, H, Hanaoka, H, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ubiquitin-interacting Motif of S5a Bound to the Ubiquitin-like Domain of HR23B
J.Biol.Chem., 279, 2004
|
|
1U3Z
 
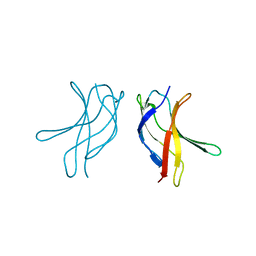 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U98
 
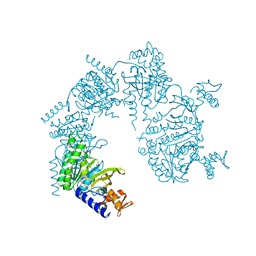 | |
4EP4
 
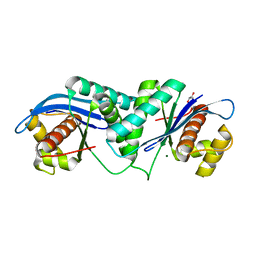 | | Thermus thermophilus RuvC structure | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, GLYCEROL, MAGNESIUM ION | | Authors: | Chen, L, Shi, K, Yin, Z.Q, Aihara, H. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution.
Nucleic Acids Res., 41, 2013
|
|
1U3J
 
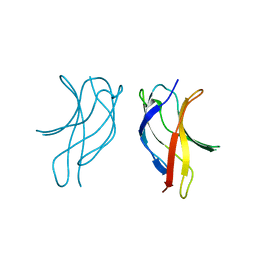 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
3WRW
 
 | |
4D94
 
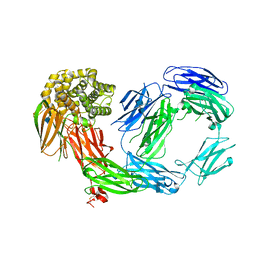 | | Crystal Structure of TEP1r | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Le, B.V, Williams, M, Logarajah, S, Baxter, R.H.G. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis for Genetic Resistance of Anopheles gambiae to Plasmodium: Structural Analysis of TEP1 Susceptible and Resistant Alleles.
Plos Pathog., 8, 2012
|
|
1UII
 
 | |
1ULK
 
 | | Crystal Structure of Pokeweed Lectin-C | | Descriptor: | lectin-C | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
1ULM
 
 | | Crystal Structure of Pokeweed Lectin-D2 complexed with tri-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin-D2 | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
4DII
 
 | | X-ray structure of the complex between human alpha thrombin and thrombin binding aptamer in the presence of potassium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity.
Nucleic Acids Res., 40, 2012
|
|
1V3V
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complexed with NADP and 15-oxo-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
