8FR9
 
 | |
8FRG
 
 | |
6YIN
 
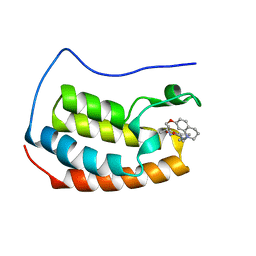 | | Crystal structure of the first bromodomain of BRD4 in complex with a benzo-diazepine ligand | | Descriptor: | (4~{R})-~{N}-[3-(7-methoxy-3,4-dihydro-2~{H}-quinolin-1-yl)propyl]-4-methyl-2-oxidanylidene-1,3,4,5-tetrahydro-1,5-benzodiazepine-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of the first bromodomain of BRD4 in complex with a benzo-diazepine ligand
To Be Published
|
|
6N13
 
 | | UbcH7-Ub Complex with R0RBR Parkin and phosphoubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Condos, T.E.C, Dunkerley, K.M, Freeman, E.A, Barber, K.R, Aguirre, J.D, Chaugule, V.K, Xiao, Y, Konermann, L, Walden, H, Shaw, G.S. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
EMBO J., 37, 2018
|
|
8D3S
 
 | | HIV-1 Integrase Catalytic Core Domain F185H Mutant Complexed with BKC-110 | | Descriptor: | (2S)-tert-butoxy{4-(4-chlorophenyl)-2,6-dimethyl-1-[(1-methyl-1H-pyrazol-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridin-5-yl}acetic acid, Integrase | | Authors: | Dinh, T, Kvaratskhelia, M. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structural and mechanistic bases for the viral resistance to allosteric HIV-1 integrase inhibitor pirmitegravir.
Biorxiv, 2024
|
|
1O8S
 
 | | Structure of CsCBM6-3 from Clostridium stercorarium in complex with cellobiose | | Descriptor: | CALCIUM ION, PUTATIVE ENDO-XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilbrun, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
3NYS
 
 | |
3QQV
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from corynebacterium glutamicum complexed with isoprenyl diphosphate and magnesium | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Poulter, C.D, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7JDW
 
 | |
8UH8
 
 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | Descriptor: | ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-01-24 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
8GAG
 
 | | Cannabinoid receptor 1-Gi complex with novel ligand | | Descriptor: | Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tummino, T.A, Iliopoulos-Tsoutsouvas, C, Braz, J.M, O'Brien, E.S, Krishna Kumar, K, Makriyannis, M, Basbaum, A.I, Shoichet, B.K. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cannabinoid receptor 1-Gi complex with novel ligand
To Be Published
|
|
5HG0
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM | | Descriptor: | Pantothenate synthetase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM
To Be Published
|
|
4APB
 
 | | Crystal structure of Mycobacterium tuberculosis fumarase (Rv1098c) S318C in complex with fumarate | | Descriptor: | CALCIUM ION, FUMARATE HYDRATASE CLASS II, FUMARIC ACID | | Authors: | Bellinzoni, M, Haouz, A, Mechaly, A.E, Alzari, P.M. | | Deposit date: | 2012-03-31 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
6YIL
 
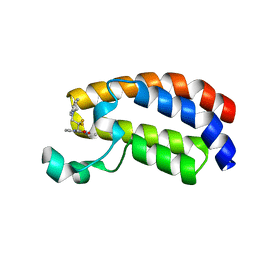 | | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand | | Descriptor: | (3~{R})-~{N}-[3-(3,4-dihydro-2~{H}-quinolin-1-yl)-2,2-bis(fluoranyl)propyl]-3-methyl-2-oxidanylidene-3,4-dihydro-1~{H}-quinoxaline-5-carboxamide, CREBBP | | Authors: | Picaud, S, Brand, M, Tobias, K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Conway, S, Filippakopoulos, P. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal structure of the CREBBP bromodomain in complex with a tetrahydroquinoxaline ligand
To Be Published
|
|
3OXE
 
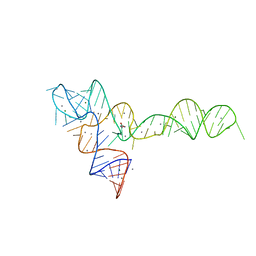 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
7JJH
 
 | |
8I5I
 
 | |
8I5H
 
 | |
2G2P
 
 | | Crystal Structure of E.coli transthyretin-related protein with bound Zn and Br | | Descriptor: | BROMIDE ION, SULFATE ION, Transthyretin-like protein, ... | | Authors: | Lundberg, E, Backstrom, S, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2006-02-16 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The transthyretin-related protein: structural investigation of a novel protein family
J.Struct.Biol., 155, 2006
|
|
8UH5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-06 | | Release date: | 2023-12-13 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
8UH9
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-20 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
1XMM
 
 | | Structure of human Dcps bound to m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, N7-METHYL-GUANOSINE-5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Chen, N, Song, H. | | Deposit date: | 2004-10-04 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human DcpS in ligand-free and m7GDP-bound forms suggest a dynamic mechanism for scavenger mRNA decapping.
J.Mol.Biol., 347, 2005
|
|
1CUA
 
 | | CUTINASE, N172K MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1ZB5
 
 | | Recognition of peptide ligands by signalling protein from porcine mammary gland (SPP-40): Crystal structure of the complex of SPP-40 with a peptide Trp-Pro-Trp at 2.45A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PEPTIDE TRP-PRO-TRP, signal processing protein | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Somvanshi, R.K, Sharma, S, Dey, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-04-07 | | Release date: | 2005-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Recognition of peptide ligands by signalling protein from porcine mammary gland (SPP-40): Crystal structure of the complex of SPP-40 with a peptide Trp-Pro-Trp at 2.45A resolution
To be Published
|
|
2G2N
 
 | | Crystal Structure of E.coli transthyretin-related protein with bound Zn | | Descriptor: | SULFATE ION, Transthyretin-like protein, ZINC ION | | Authors: | Lundberg, E, Backstrom, S, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2006-02-16 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The transthyretin-related protein: structural investigation of a novel protein family
J.Struct.Biol., 155, 2006
|
|
