1L6R
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | Descriptor: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-03-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
2HQN
 
 | |
1A1X
 
 | | CRYSTAL STRUCTURE OF MTCP-1 INVOLVED IN T CELL MALIGNANCIES | | Descriptor: | HMTCP-1 | | Authors: | Fu, Z.Q, Dubois, G.C, Song, S.P, Kulikovskaya, I, Virgilio, L, Rothstein, J, Croce, C.M, Weber, I.T, Harrison, R.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MTCP-1: implications for role of TCL-1 and MTCP-1 in T cell malignancies.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3PPT
 
 | | REP1-NXSQ fatty acid transporter | | Descriptor: | PALMITOLEIC ACID, ReP1-NCXSQ | | Authors: | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | Deposit date: | 2010-11-25 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5OQ0
 
 | |
5X67
 
 | | Human thymidylate synthase in complex with dUMP and nolatrexed | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-azanyl-6-methyl-5-pyridin-4-ylsulfanyl-3H-quinazolin-4-one, Thymidylate synthase | | Authors: | Chen, D, Jansson, A, Larsson, A, Nordlund, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural analyses of human thymidylate synthase reveal a site that may control conformational switching between active and inactive states.
J. Biol. Chem., 292, 2017
|
|
1IHI
 
 | | Crystal Structure of Human Type III 3-alpha-Hydroxysteroid Dehydrogenase/Bile Acid Binding Protein (AKR1C2) Complexed with NADP+ and Ursodeoxycholate | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE, ISO-URSODEOXYCHOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jin, Y, Stayrook, S.E, Albert, R.H, Palackal, N.T, Penning, T.M, Lewis, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human type III 3alpha-hydroxysteroid dehydrogenase/bile acid binding protein complexed with NADP(+) and ursodeoxycholate.
Biochemistry, 40, 2001
|
|
1V0Y
 
 | |
3PP6
 
 | | REP1-NXSQ fatty acid transporter Y128F mutant | | Descriptor: | PALMITOLEIC ACID, ReP1-NCXSQ | | Authors: | Berberian, G, Bollo, M, Howard, E, Cousido-Siah, A, Mitschler, A, Ayoub, D, Sanglier-Cianferani, S, Van Dorsselaer, A, DiPolo, R, Beauge, L, Petrova, T, Schulze-Briese, C, Wang, M, Podjarny, A. | | Deposit date: | 2010-11-24 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional studies of ReP1-NCXSQ, a protein regulating the squid nerve Na+/Ca2+ exchanger.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5XLK
 
 | | Crystal structure of the flagellar cap protein FliD D2-D3 domains from Serratia marcescens in Space group I422 | | Descriptor: | Flagellar hook-associated protein 2, ZINC ION | | Authors: | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
1J2U
 
 | | Creatininase Zn | | Descriptor: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
3L5X
 
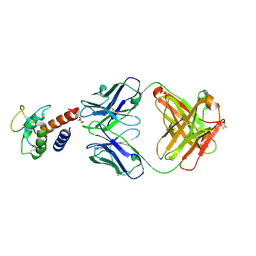 | | Crystal structure of the complex between IL-13 and H2L6 FAB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, H2L6 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
2HVV
 
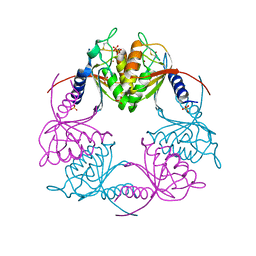 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | Descriptor: | SULFATE ION, ZINC ION, deoxycytidylate deaminase | | Authors: | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
2HVW
 
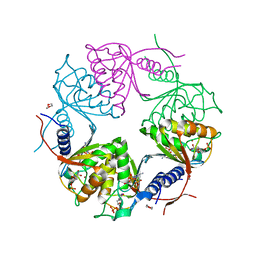 | | Crystal structure of dCMP deaminase from Streptococcus mutans | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 3,4-DIHYDRO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Hou, H.F, Gao, Z.Q, Li, L.F, Liang, Y.H, Su, X.D, Dong, Y.H. | | Deposit date: | 2006-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of Streptococcus mutans 2'-deoxycytidylate deaminase and its complex with substrate analog and allosteric regulator dCTP x Mg2+.
J.Mol.Biol., 377, 2008
|
|
3LTM
 
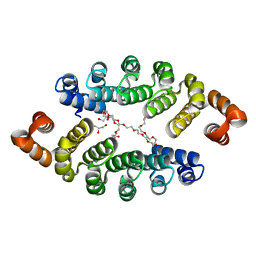 | | Structure of a new family of artificial alpha helicoidal repeat proteins (alpha-Rep) based on thermostable HEAT-like repeats | | Descriptor: | Alpha-Rep4, DODECAETHYLENE GLYCOL, GLYCEROL, ... | | Authors: | Urvoas, A, Guellouz, A, Graille, M, van Tilbeurgh, H, Desmadril, M, Minard, P. | | Deposit date: | 2010-02-16 | | Release date: | 2010-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design, production and molecular structure of a new family of artificial alpha-helicoidal repeat proteins ( alpha Rep) based on thermostable HEAT-like repeats
J.Mol.Biol., 404, 2010
|
|
3L5W
 
 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
6ALY
 
 | | Solution structure of yeast Med15 ABD2 residues 277-368 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 15 | | Authors: | Tuttle, L.M, Pacheco, D, Warfield, L, Hahn, S, Klevit, R.E. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gcn4-Mediator Specificity Is Mediated by a Large and Dynamic Fuzzy Protein-Protein Complex.
Cell Rep, 22, 2018
|
|
3L7F
 
 | | Structure of IL-13 antibody H2L6, A humanized variant OF C836 | | Descriptor: | CALCIUM ION, H2L6 HEAVY CHAIN, H2L6 LIGHT CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
1V0S
 
 | |
1V0W
 
 | |
1V0T
 
 | |
1J2T
 
 | | Creatininase Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, ZINC ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
1M6V
 
 | | Crystal Structure of the G359F (small subunit) Point Mutant of Carbamoyl Phosphate Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-07-17 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl-phosphate synthetase. Creation of an escape route for ammonia
J.Biol.Chem., 277, 2002
|
|
1AUW
 
 | | H91N DELTA 2 CRYSTALLIN FROM DUCK | | Descriptor: | DELTA 2 CRYSTALLIN | | Authors: | Abu-Abed, M, Vallee, F, Howell, P.L. | | Deposit date: | 1997-09-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of the enzymatically active and inactive forms of delta crystallin and the role of histidine 91.
Biochemistry, 36, 1997
|
|
