6O9A
 
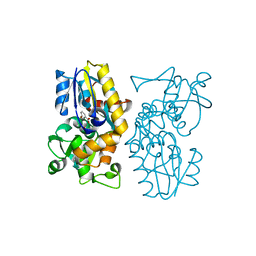 | | Crystal structure of MqnA complexed with 3-hydroxybenzoic acid | | Descriptor: | 3-HYDROXYBENZOIC ACID, ACETATE ION, Chorismate dehydratase | | Authors: | Hicks, K.A, Mahanta, N, Naseem, S, Fedoseyenko, D, Begley, T.P, Ealick, S.E. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.326 Å) | | Cite: | Menaquinone Biosynthesis: Biochemical and Structural Studies of Chorismate Dehydratase.
Biochemistry, 58, 2019
|
|
6OBO
 
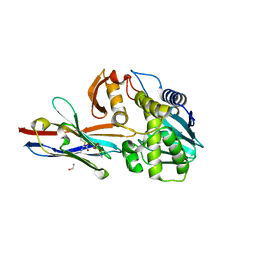 | | Ricin A chain bound to VHH antibody V6A6 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intracellular Neutralization of Ricin Toxin by Single-domain Antibodies Targeting the Active Site.
J.Mol.Biol., 432, 2020
|
|
3CBX
 
 | |
6OC7
 
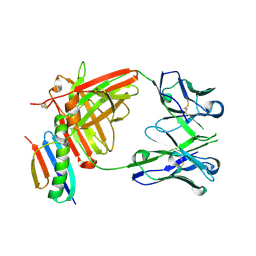 | | HMP42 Fab in complex with Protein G | | Descriptor: | Heavy chain of HMP42 Fab, Immunoglobulin G-binding protein G, Light chain for HMP42 Fab | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2019-03-22 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
3CC8
 
 | |
6O9L
 
 | | Human holo-PIC in the closed state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OCH
 
 | | Crystal structure of VASH1-SVBP complex bound with parthenolide | | Descriptor: | GLYCEROL, SULFATE ION, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3CCW
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[4-(benzylcarbamoyl)-2-(4-fluorophenyl)-5-(1-methylethyl)-1H-imidazol-1-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
6OCX
 
 | |
6ODA
 
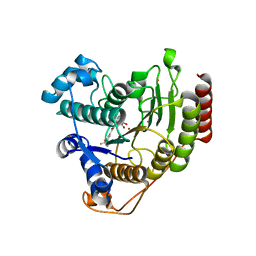 | | Crystal structure of HDAC8 in complex with compound 2 | | Descriptor: | Histone deacetylase 8, N-{2-[3-(hydroxyamino)-3-oxopropyl]phenyl}-3-(trifluoromethyl)benzamide, POTASSIUM ION, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
3CD3
 
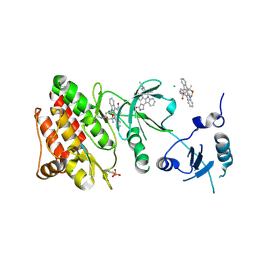 | | Crystal structure of phosphorylated human feline sarcoma viral oncogene homologue (v-FES) in complex with staurosporine and a consensus peptide | | Descriptor: | CHLORIDE ION, Proto-oncogene tyrosine-protein kinase Fes/Fps, STAUROSPORINE, ... | | Authors: | Filippakopoulos, P, Salah, E, Cooper, C, Picaud, S.S, Elkins, J.M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Coupling of SH2-Kinase Domains Links Fes and Abl Substrate Recognition and Kinase Activation
Cell(Cambridge,Mass.), 134, 2008
|
|
6OE1
 
 | |
6OE5
 
 | |
6OAW
 
 | | Crystal structure of a CRISPR Cas-related protein | | Descriptor: | UNKNOWN ATOM OR ION, WYL1 | | Authors: | Zhang, H, Dong, C, Li, L, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-18 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the modulatory role of the accessory protein WYL1 in the Type VI-D CRISPR-Cas system.
Nucleic Acids Res., 47, 2019
|
|
6OB5
 
 | | Computationally-designed, modular sense/response system (S3-2D) | | Descriptor: | Ankyrin Repeat Domain (AR), S3-2D variant, FARNESYL DIPHOSPHATE, ... | | Authors: | Thompson, M.C, Glasgow, A.A, Huang, Y.M, Fraser, J.S, Kortemme, T. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.208 Å) | | Cite: | Computational design of a modular protein sense-response system.
Science, 366, 2019
|
|
3K6N
 
 | | Crystal structure of the S225E mutant Kir3.1 cytoplasmic pore domain | | Descriptor: | G protein-activated inward rectifier potassium channel 1, SODIUM ION | | Authors: | Xu, Y, Shin, H.G, Szep, S, Lu, Z. | | Deposit date: | 2009-10-09 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Physical determinants of strong voltage sensitivity of K(+) channel block.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6OEJ
 
 | |
6OEV
 
 | | Structure of human Patched1 in complex with native Sonic Hedgehog | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-17 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of human Patched and its complex with native palmitoylated sonic hedgehog.
Nature, 560, 2018
|
|
3K6U
 
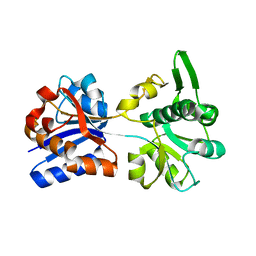 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6OBL
 
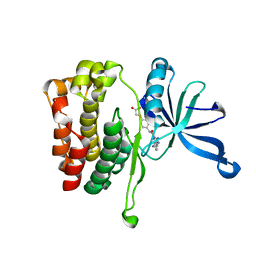 | | JAK2 JH2 in complex with JAK168 | | Descriptor: | Tyrosine-protein kinase JAK2, [4-({5-amino-3-[(4-cyanophenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenoxy]acetic acid | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6OBR
 
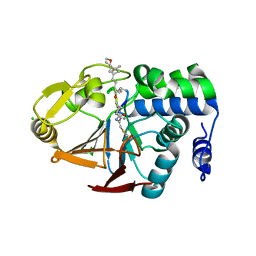 | | PP1 Y134A in complex with Microcystin LR | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Microcystin LR, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3C9N
 
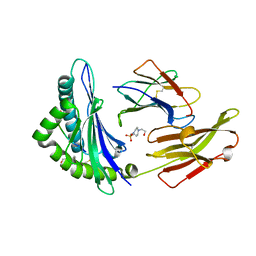 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
6OCB
 
 | |
3CAF
 
 | |
6OCR
 
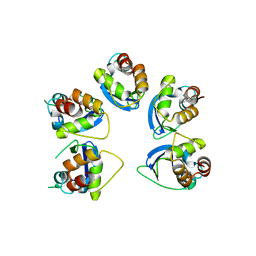 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
