3EG3
 
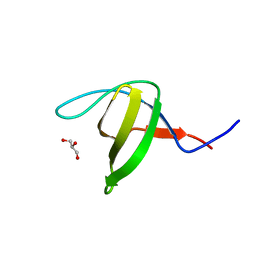 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EG0
 
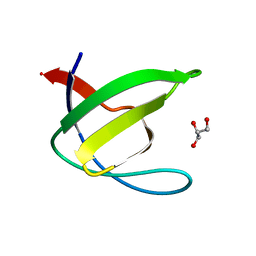 | | Crystal structure of the N114T mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
6BGC
 
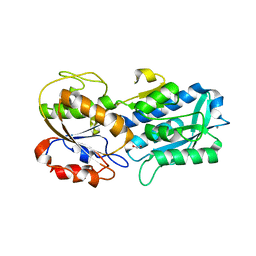 | | The crystal structure of the W145A variant of TpMglB-2 (Tp0684) with bound glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/galactose-binding lipoprotein, ... | | Authors: | Brautigam, C.A, Norgard, M.V, Deka, R.K. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Crystal structures of MglB-2 (TP0684), a topologically variant d-glucose-binding protein from Treponema pallidum, reveal a ligand-induced conformational change.
Protein Sci., 27, 2018
|
|
7EWE
 
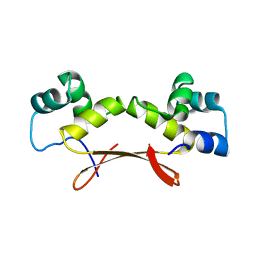 | | Mycobacterium tuberculosis HigA2 (Form III) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
7EWC
 
 | | Mycobacterium tuberculosis HigA2 (Form I) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
7EWD
 
 | | Mycobacterium tuberculosis HigA2 (Form II) | | Descriptor: | Putative antitoxin HigA2 | | Authors: | Kim, H.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chasing the structural diversity of the transcription regulator Mycobacterium tuberculosis HigA2.
Iucrj, 8, 2021
|
|
6IRP
 
 | |
6CF1
 
 | | Proteus vulgaris HigA antitoxin structure | | Descriptor: | Antitoxin HigA, POTASSIUM ION | | Authors: | Schureck, M.A, Hoffer, E.D, Ei Cho, S, Dunham, C.M. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
6CHV
 
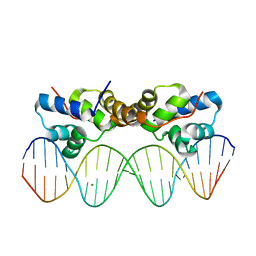 | | Proteus vulgaris HigA antitoxin bound to DNA | | Descriptor: | Antitoxin HigA, MAGNESIUM ION, pHigCryst3, ... | | Authors: | Schureck, M.A, Hoffer, E.D, Onuoha, N, Dunham, C.M. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
1BOS
 
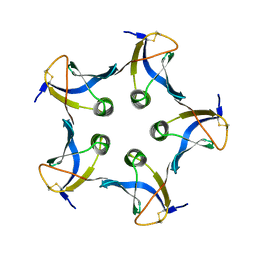 | | SHIGA-LIKE TOXIN COMPLEXED WITH ITS RECEPTOR | | Descriptor: | SHIGA-LIKE TOXIN I B SUBUNIT, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Ling, H, Boodhoo, A, Hazes, B, Cummings, M.D, Armstrong, G.D, Brunton, J.L, Read, R.J. | | Deposit date: | 1998-01-13 | | Release date: | 1999-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3.
Biochemistry, 37, 1998
|
|
7KDQ
 
 | |
8SKV
 
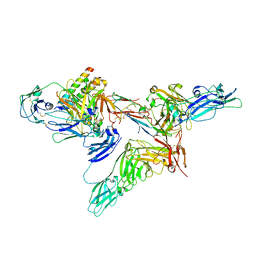 | |
6OH1
 
 | | IgA1 Protease G5 domain structure | | Descriptor: | Immunoglobulin A1 protease | | Authors: | Eisenmesser, E.Z, Chi, Y.C, Paukovich, N, Redzic, J.S, Rahkola, J.T, Janoff, E.N. | | Deposit date: | 2019-04-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Streptococcus pneumoniae G5 domains bind different ligands.
Protein Sci., 28, 2019
|
|
1KXO
 
 | | ENGINEERED LIPOCALIN DIGA16 : APO-FORM | | Descriptor: | DigA16 | | Authors: | Korndoerfer, I.P, Skerra, A. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural mechanism of specific ligand recognition by a lipocalin tailored for the complexation of digoxigenin.
J.Mol.Biol., 330, 2003
|
|
6XJB
 
 | | IgA1 Protease | | Descriptor: | Immunoglobulin A1 protease | | Authors: | Eisenmesser, E.Z, Zheng, H. | | Deposit date: | 2020-06-23 | | Release date: | 2020-12-09 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism and inhibition of Streptococcus pneumoniae IgA1 protease.
Nat Commun, 11, 2020
|
|
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
7TO5
 
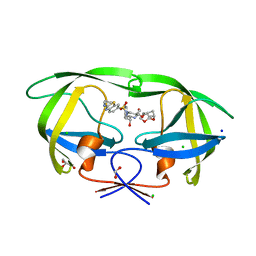 | | HIV-1 wild type protease with GRL-05816A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 1] | | Descriptor: | (1R,3aS,4S,6S,7aR)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
7TO6
 
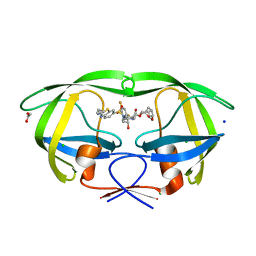 | | HIV-1 wild type protease with GRL-01717A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 2] | | Descriptor: | (1S,3aR,4R,6R,7aS)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
2QTJ
 
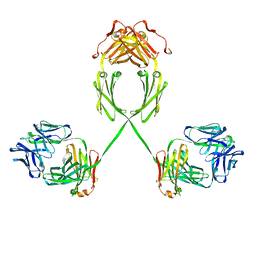 | | Solution structure of human dimeric immunoglobulin A | | Descriptor: | Ig alpha-1 chain C region, Kappa light chain IgA1 | | Authors: | Bonner, A, Furtado, P.B, Almogren, A, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Implications of the near-planar solution structure of human myeloma dimeric IgA1 for mucosal immunity and IgA nephropathy
J.Immunol., 180, 2008
|
|
6Q2S
 
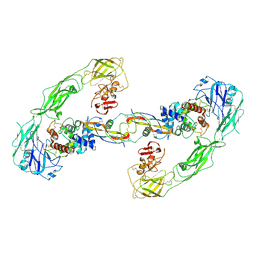 | | Cryo-EM structure of RET/GFRa3/ARTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-3, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
4Z41
 
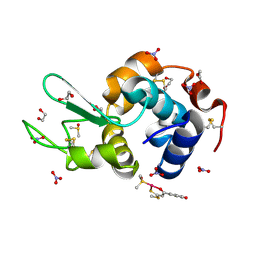 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Compound with a S,O Bidentate Ligand (9a=Chloro-(1-(3'-hydroxy)-3-(methylthio)-3-thioxo-prop-1-en-1-olate-O,S)-(dimethylsulfoxide-S)-platinum(II)) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-04-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
4Z3M
 
 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Complex with S,O Bidentate Ligands (9b) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-03-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
1AI6
 
 | | PENICILLIN ACYLASE WITH P-HYDROXYPHENYLACETIC ACID | | Descriptor: | 4-HYDROXYPHENYLACETATE, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AI7
 
 | | PENICILLIN ACYLASE COMPLEXED WITH PHENOL | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, PHENOL | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
6Q2R
 
 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex in the tetrameric form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
