4HGJ
 
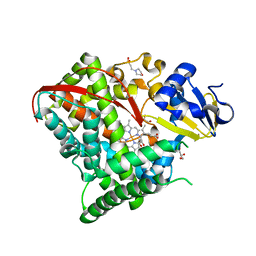 | | Crystal structure of P450 BM3 5F5 heme domain variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4N96
 
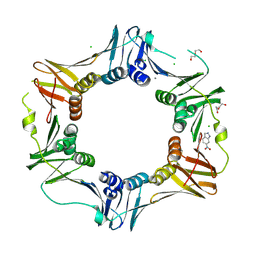 | | E. coli sliding clamp in complex with 6-nitroindazole | | Descriptor: | 6-NITROINDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4OFS
 
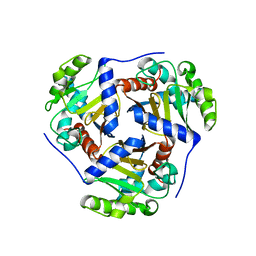 | | Crystal structure of a truncated catalytic core of the 2-oxoacid dehydrogenase multienzyme complex from Thermoplasma acidophilum | | Descriptor: | Probable lipoamide acyltransferase | | Authors: | Marrot, N.L, Marshall, J.J.T, Svergun, D.I, Crennell, S.J, Hough, D.W, van den Elsen, J.M.H, Danson, M.J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex.
Biochem.J., 463, 2014
|
|
4HGG
 
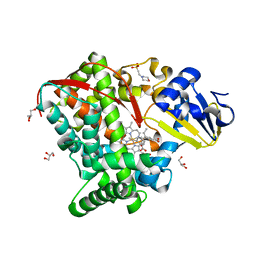 | | Crystal structure of P450 BM3 5F5R heme domain variant complexed with styrene | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGI
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset II) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4N9A
 
 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N97
 
 | | E. coli sliding clamp in complex with 5-nitroindole | | Descriptor: | 5-nitro-1H-indole, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
5VPL
 
 | | CRYSTAL STRUCTURE OF DER F 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C1 - HEAVY CHAIN, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
4NIP
 
 | |
4NHQ
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (5 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NIN
 
 | |
4L8C
 
 | | Crystal structure of the H2Db in complex with the NP-N3D peptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Rossjohn, J, Gras, S. | | Deposit date: | 2013-06-16 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acute emergence and reversion of influenza A virus quasispecies within CD8(+) T cell antigenic peptides.
Nat Commun, 4, 2013
|
|
4FTH
 
 | | Crystal Structure of NtrC4 DNA-binding domain bound to double-stranded DNA | | Descriptor: | 5'-D(*AP*CP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*GP*CP*AP*T)-3', 5'-D(P*GP*AP*TP*GP*CP*AP*TP*TP*TP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*AP*A)-3', Transcriptional regulator (NtrC family) | | Authors: | Vidangos, N.K, Heideker, J, Lyubimov, A.Y, Lamers, M, Huo, Y, Pelton, J.G, Ton, J, Gralla, J.D, Kuriyan, J, Berger, J.M, Wemmer, D.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | DNA Recognition by a sigma (54) Transcriptional Activator from Aquifex aeolicus.
J.Mol.Biol., 426, 2014
|
|
7DWS
 
 | |
4D48
 
 | | Crystal Structure of glucose-1-phosphate uridylyltransferase GalU from Erwinia amylovora. | | Descriptor: | GLUCOSE-1-PHOSPHATE URIDYLYLTRANSFERASE | | Authors: | Toccafondi, M, Wuerges, J, Cianci, M, Benini, S. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Glucose-1-phosphate uridylyltransferase from Erwinia amylovora: Activity, structure and substrate specificity.
Biochim. Biophys. Acta, 1865, 2017
|
|
4COQ
 
 | | The complex of alpha-Carbonic anhydrase from Thermovibrio ammonificans with inhibitor sulfanilamide. | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CARBONATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J.A. | | Deposit date: | 2014-01-30 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6M1V
 
 | |
4E2K
 
 | | The structure of the S. aureus DnaG RNA Polymerase Domain | | Descriptor: | BENZAMIDINE, DNA primase | | Authors: | Rymer, R.U, Solorio, F.A, Clement, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-08 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4DCB
 
 | | Y. pestis Plasminogen Activator Pla in Complex with Human Plasminogen Activation Loop Peptide ALP11 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ... | | Authors: | Eren, E, van den Berg, B. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Structural basis for activation of an integral membrane protease by lipopolysaccharide.
J.Biol.Chem., 287, 2012
|
|
4DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH LEU 33 REPLACED BY THR, THR 34 REPLACED BY GLY, ASP 36 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1997-01-06 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
4HKQ
 
 | | XMRV reverse transcriptase in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*TP*GP*GP*AP*AP*TP*CP*A*GP*GP*TP*GP*TP*CP*GP*CP*AP*CP*TP*CP*TP*G)-3'), RNA (5'-R(*AP*AP*CP*AP*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*CP*AP*U)-3'), Reverse transcriptase/ribonuclease H p80 | | Authors: | Nowak, E, Potrzebowski, W, Konarev, P.V, Rausch, J.W, Bona, M.K, Svergun, D.I, Bujnicki, J.M, Le Grice, S.F.J, Nowotny, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural analysis of monomeric retroviral reverse transcriptase in complex with an RNA/DNA hybrid
Nucleic Acids Res., 41, 2013
|
|
7EKE
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKC
 
 | | Structure of SARS-CoV-2 Gamma variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
