5GJ4
 
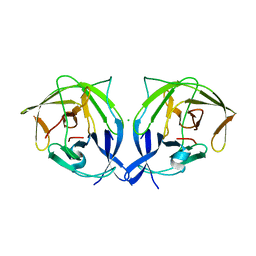 | |
1B03
 
 | |
1CWW
 
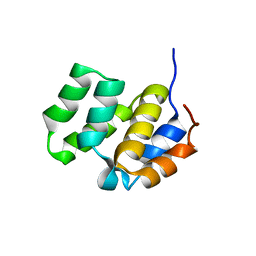 | | SOLUTION STRUCTURE OF THE CASPASE RECRUITMENT DOMAIN (CARD) FROM APAF-1 | | Descriptor: | APOPTOTIC PROTEASE ACTIVATING FACTOR 1 | | Authors: | Day, C.L, Dupont, C, Lackmann, M, Vaux, D.L, Hinds, M.G. | | Deposit date: | 1999-08-26 | | Release date: | 2000-01-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and mutagenesis of the caspase recruitment domain (CARD) from Apaf-1.
Cell Death Differ., 6, 1999
|
|
1DL0
 
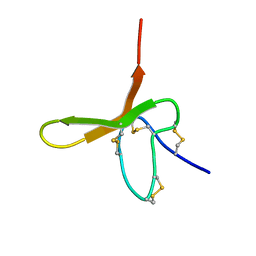 | |
5U5S
 
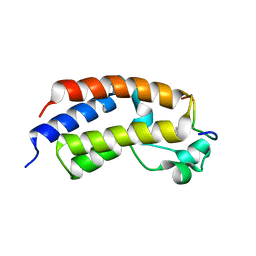 | |
5TR5
 
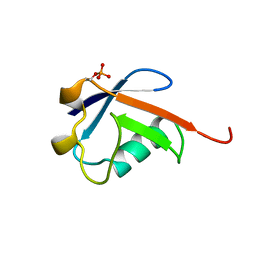 | |
5V2R
 
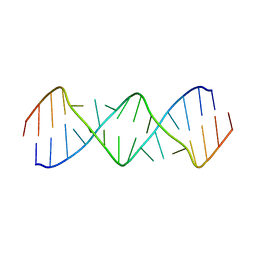 | | Structure of a GA Rich 8x8 Nucleotide RNA Internal Loop | | Descriptor: | RNA (5'-R(*CP*CP*AP*GP*AP*AP*AP*CP*GP*GP*AP*UP*GP*GP*A)-3') | | Authors: | Kauffmann, A.D, Kennedy, S.D, Turner, D.H. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure of an 8 8 Nucleotide RNA Internal Loop Flanked on Each Side by Three Watson-Crick Pairs and Comparison to Three-Dimensional Predictions.
Biochemistry, 56, 2017
|
|
5UZF
 
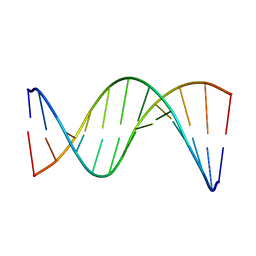 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNA structure | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*AP*AP*AP*AP*TP*CP*G)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
5UZD
 
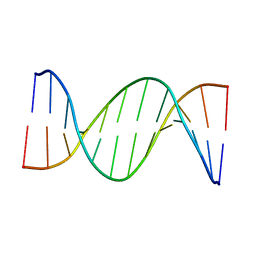 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A2-DNA structure | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*GP*AP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*AP*TP*CP*GP*AP*TP*GP*C)-3') | | Authors: | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | Deposit date: | 2017-02-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6RSA
 
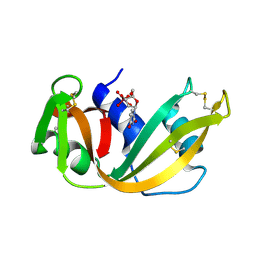 | |
6TOB
 
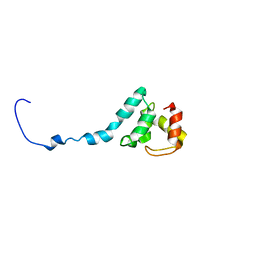 | |
6GWX
 
 | | Stabilising and Understanding a Miniprotein by Rational Design. | | Descriptor: | Optimised PPa-TYR | | Authors: | Porter Goff, K.L, Williams, C, Baker, E.G, Nicol, D, Samphire, J.L, Zieleniewski, F.L, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Stabilizing and Understanding a Miniprotein by Rational Redesign.
Biochemistry, 58, 2019
|
|
1JM7
 
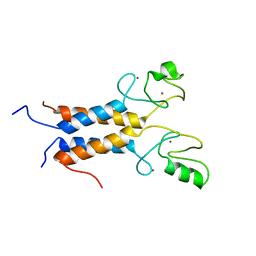 | | Solution structure of the BRCA1/BARD1 RING-domain heterodimer | | Descriptor: | BRCA1-ASSOCIATED RING DOMAIN PROTEIN 1, BREAST CANCER TYPE 1 SUSCEPTIBILITY PROTEIN, ZINC ION | | Authors: | Brzovic, P.S, Rajagopal, P, Hoyt, D.W, King, M.-C, Klevit, R.E. | | Deposit date: | 2001-07-17 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a BRCA1-BARD1 heterodimeric RING-RING complex.
Nat.Struct.Biol., 8, 2001
|
|
5VMU
 
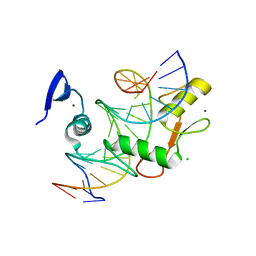 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a double CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*(5CM)P*GP*(5CM)P*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
5VMZ
 
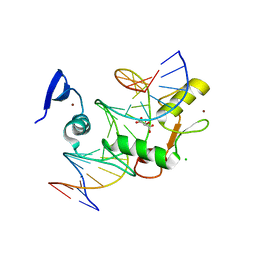 | | Kaiso (ZBTB33) E535Q mutant zinc finger DNA binding domain in complex with a double CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*(5CM)P*GP*(5CM)P*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
7WE3
 
 | |
7WEI
 
 | |
5J8H
 
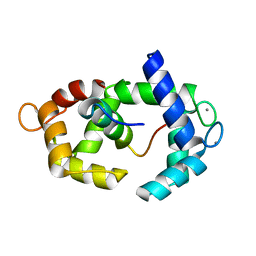 | | Structure of calmodulin in a complex with a peptide derived from a calmodulin-dependent kinase | | Descriptor: | CALCIUM ION, Calmodulin, Eukaryotic elongation factor 2 kinase | | Authors: | Alphonse, S, Lee, K, Piserchio, A, Tavares, C.D.J, Giles, D.H, Wellmann, R.M, Dalby, K.N, Ghose, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Eukaryotic Elongation Factor 2 Kinase by Calmodulin.
Structure, 24, 2016
|
|
5B88
 
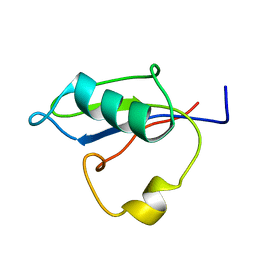 | | RRM-like domain of DEAD-box protein, CsdA | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Peng, J, Zhang, J, Wu, J, Tang, Y, Shi, Y. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
4B8T
 
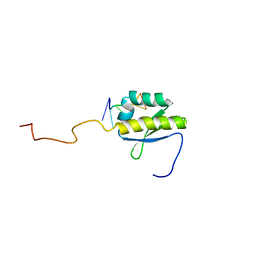 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
7X89
 
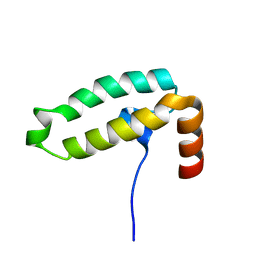 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
3OL3
 
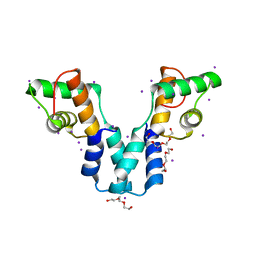 | |
3OL4
 
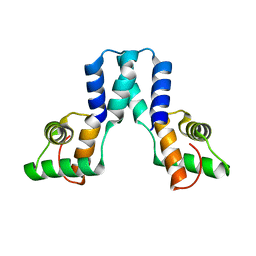 | |
3OV5
 
 | |
