2ZJL
 
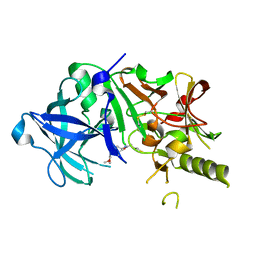 | | Crystal structure of the human BACE1 catalytic domain in complex with N-[1-(5-bromo-2,3-dimethoxy-benzyl)-piperidin-4-yl]-4-mercapto-butyramide | | Descriptor: | Beta-secretase 1, N-[1-(5-bromo-2,3-dimethoxybenzyl)piperidin-4-yl]-4-sulfanylbutanamide | | Authors: | Randal, M, Lam, M.B, Lu, W, Romanowski, M.J. | | Deposit date: | 2008-03-07 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-based discovery of novel BACE1 inhibitors using Tethering technology
To be Published
|
|
1POS
 
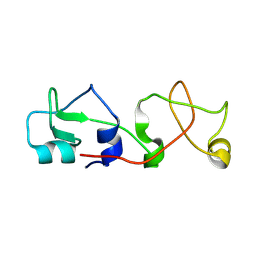 | | CRYSTAL STRUCTURE OF A NOVEL DISULFIDE-LINKED "TREFOIL" MOTIF FOUND IN A LARGE FAMILY OF PUTATIVE GROWTH FACTORS | | Descriptor: | PORCINE PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | De, A, Brown, D, Gorman, M, Carr, M, Sanderson, M.R, Freemont, P.S. | | Deposit date: | 1993-10-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a disulfide-linked "trefoil" motif found in a large family of putative growth factors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
2M2V
 
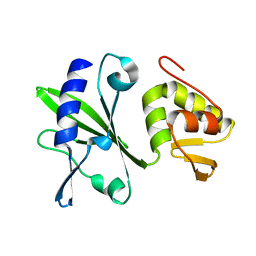 | |
1PSD
 
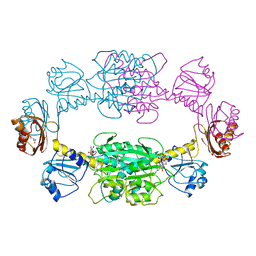 | |
2PUV
 
 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 5-AMINO-5-DEOXY-1-O-PHOSPHONO-D-MANNITOL, ACETATE ION, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
2HKE
 
 | | Mevalonate diphosphate decarboxylase from Trypanosoma brucei | | Descriptor: | Diphosphomevalonate decarboxylase, putative, SULFATE ION | | Authors: | Byres, E, Alphey, M.S, Hunter, W.N. | | Deposit date: | 2006-07-04 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Trypanosoma brucei and Staphylococcus aureus Mevalonate Diphosphate Decarboxylase Inform on the Determinants of Specificity and Reactivity
J.Mol.Biol., 371, 2007
|
|
2PUT
 
 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | ACETATE ION, FRUCTOSE -6-PHOSPHATE, SODIUM ION, ... | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
1WZE
 
 | |
4HSR
 
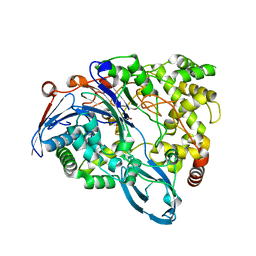 | | Crystal Structure of a class III engineered cephalosporin acylase | | Descriptor: | 5,5-dihydroxy-L-norvaline, glutaryl-7-aminocephalosporanic acid acylase alpha chain, glutaryl-7-aminocephalosporanic acid acylase beta chain | | Authors: | Vrielink, A, Golden, E, Patterson, R, Tie, W.J, Anandan, A, Flematti, G, Molla, G, Rosini, E, Pollegioni, L. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of a class III engineered cephalosporin acylase: comparisons with class I acylase and implications for differences in substrate specificity and catalytic activity.
Biochem.J., 451, 2013
|
|
1BMD
 
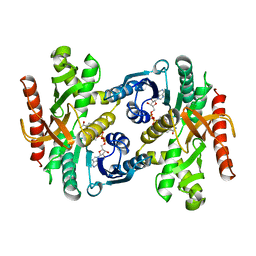 | |
7L7F
 
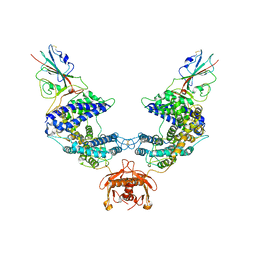 | |
6P9Y
 
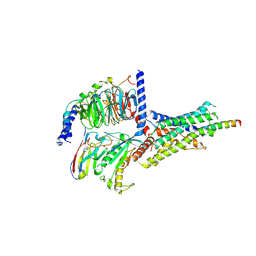 | | PAC1 GPCR Receptor complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-06-10 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation.
Mol.Cell, 77, 2020
|
|
7L7K
 
 | |
5MOA
 
 | | ABA RECEPTOR FROM TOMATO, SlPYL1 | | Descriptor: | SlPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MTQ
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT511 | | Descriptor: | 2-[4-[(4-cyclohexyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5MMQ
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | CSPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MN0
 
 | | ABA RECEPTOR FROM CITRUS, CSPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, CHLORIDE ION, CSPYL1, ... | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MH6
 
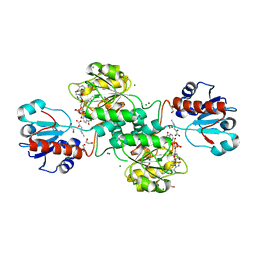 | | D-2-hydroxyacid dehydrogenases (D2-HDH) from Haloferax mediterranei in complex with 2-ketohexanoic acid and NAD+ (1.35 A resolution) | | Descriptor: | 1,2-ETHANEDIOL, 2-Ketohexanoic acid, D-2-hydroxyacid dehydrogenase, ... | | Authors: | Bisson, C, Baker, P.J, Domenech Perez, J, Pramanpol, N, Harding, S.E, Rice, D.W, Ferrer, J. | | Deposit date: | 2016-11-23 | | Release date: | 2018-06-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Productive ternary complexes of D-2-hydroxyacid dehydrogenase provide insights into the chiral specificity of its reaction mechanism
To Be Published
|
|
4XH8
 
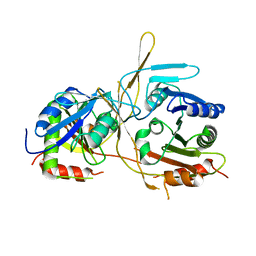 | |
5CNP
 
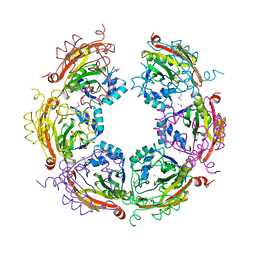 | | X-ray crystal structure of Spermidine n1-acetyltransferase from Vibrio cholerae. | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Osipiuk, J, VOLKART, L, MOY, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
5MTP
 
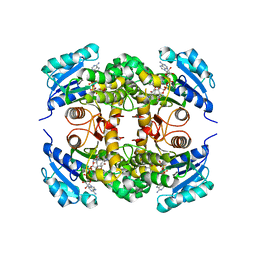 | | Crystal structure of M. tuberculosis InhA inhibited by PT514 | | Descriptor: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5C25
 
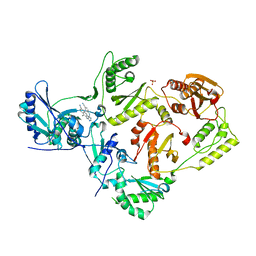 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 6-((4-((4-cyanophenyl)amino)-1,3,5-triazin-2-yl)amino)-5,7-dimethyl-2-naphthonitrile (JLJ639), a Non-nucleoside Inhibitor | | Descriptor: | 6-({4-[(4-cyanophenyl)amino]-1,3,5-triazin-2-yl}amino)-5,7- dimethyl-2-naphthonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Frey, K.M, Anderson, K.S. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Discovery and crystallography of bicyclic arylaminoazines as potent inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7KSN
 
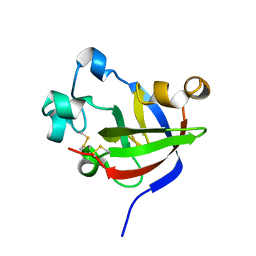 | | Crystal Structure of Sugarwin | | Descriptor: | Sugarwin | | Authors: | Maia, L.B.L, Pereira, H.M, Henrique-Silva, F, Garratt, R.C, Silva Filho, M.C. | | Deposit date: | 2020-11-23 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural and Evolutionary Analyses of PR-4 SUGARWINs Points to a Different Pattern of Protein Function
Frontiers in Plant Science, 12, 2021
|
|
5MVO
 
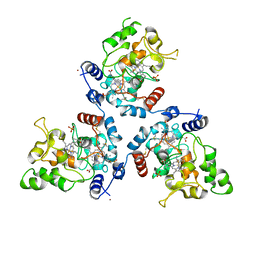 | | FoxE P43212 crystal structure of Rhodopseudomonas ferrooxidans SW2 putative iron oxidase | | Descriptor: | COPPER (II) ION, FoxE, HEME C, ... | | Authors: | Pereira, L, Saraiva, I.H, Oliveira, A.S, Soares, C, Gomes, R.O, Frazao, C. | | Deposit date: | 2017-01-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.668 Å) | | Cite: | Crystallization and preliminary crystallographic studies of FoxE from Rhodobacter ferrooxidans SW2, an Fe(II) oxidoreductase involved in photoferrotrophy.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
5CBL
 
 | |
