7CQW
 
 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
4W5N
 
 | | The Crystal Structure of Human Argonaute2 Bound to a Defined Guide RNA | | Descriptor: | MAGNESIUM ION, PHENOL, Protein argonaute-2, ... | | Authors: | Schirle, N.T, Sheu-Gruttadauria, J, MacRae, I.J. | | Deposit date: | 2014-08-18 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gene regulation. Structural basis for microRNA targeting.
Science, 346, 2014
|
|
4W6B
 
 | | Crystal Structure of a Superfolder GFP Mutant K26C Disulfide Dimer, P 21 21 21 Space Group | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, fluorescent protein K26C | | Authors: | Pashkov, I, Sawaya, M.R, Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6G
 
 | |
4W6J
 
 | | Crystal Structure of Full-Length Split GFP Mutant D117C Disulfide Dimer, P 31 2 1 Space Group | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, fluorescent protein D117C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6O
 
 | |
4W6R
 
 | |
4USG
 
 | | Crystal structure of PC4 W89Y mutant complex with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | Authors: | Zhao, Y, Liu, J. | | Deposit date: | 2014-07-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
6DR9
 
 | |
4V2I
 
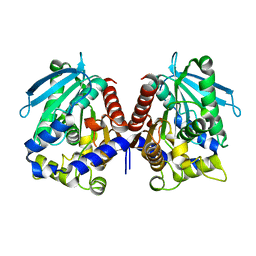 | | Biochemical characterization and structural analysis of a new cold- active and salt tolerant esterase from the marine bacterium Thalassospira sp | | Descriptor: | ESTERASE/LIPASE, MAGNESIUM ION | | Authors: | Santi, C.D, Leiros, H.-K.S, Scala, A.D, Pascale, D.D, Altermark, B, Willassen, N.-P. | | Deposit date: | 2014-10-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical Characterization and Structural Analysis of a New Cold-Active and Salt-Tolerant Esterase from the Marine Bacterium Thalassospira Sp.
Extremophiles, 20, 2016
|
|
4V74
 
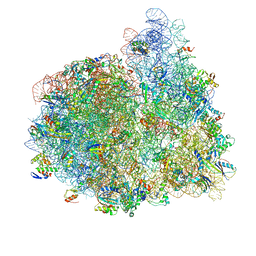 | | 70S-fMetVal-tRNAVal-tRNAfMet complex in hybrid pre-translocation state (pre5b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V7Y
 
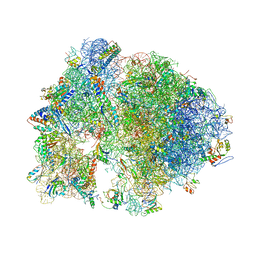 | | Structure of the Thermus thermophilus 70S ribosome complexed with azithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V70
 
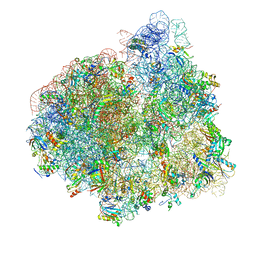 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate pre-translocation state (pre3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V7B
 
 | | Visualization of two tRNAs trapped in transit during EF-G-mediated translocation | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Ramrath, D.J.F, Lancaster, L, Sprink, T, Mielke, T, Loerke, J, Noller, H.F, Spahn, C.M.T. | | Deposit date: | 2013-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V7X
 
 | | Structure of the Thermus thermophilus ribosome complexed with erythromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-17 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V6Z
 
 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in classic pre-translocation state (pre1b) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V7Z
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with telithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5LDI
 
 | | Crystal structure of E.coli LigT in apo form | | Descriptor: | CHLORIDE ION, RNA 2',3'-cyclic phosphodiesterase | | Authors: | Myllykoski, M, Kursula, P. | | Deposit date: | 2016-06-27 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural aspects of nucleotide ligand binding by a bacterial 2H phosphoesterase.
PLoS ONE, 12, 2017
|
|
4V0L
 
 | |
4UTU
 
 | | Structural and biochemical characterization of the N- acetylmannosamine-6-phosphate 2-epimerase from Clostridium perfringens | | Descriptor: | CHLORIDE ION, N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE, N-acetylmannosamine-6-phosphate | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
2MAN
 
 | | MANNOTRIOSE COMPLEX OF THERMOMONOSPORA FUSCA BETA-MANNANASE | | Descriptor: | PROTEIN (BETA-MANNANASE), beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Hilge, M, Gloor, S.M, Piontek, K. | | Deposit date: | 1998-08-12 | | Release date: | 1999-08-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution native and complex structures of thermostable beta-mannanase from Thermomonospora fusca - substrate specificity in glycosyl hydrolase family 5.
Structure, 6, 1998
|
|
4UDR
 
 | | Crystal structure of the H467A mutant of 5-hydroxymethylfurfural oxidase (HMFO) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | Authors: | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
4WFH
 
 | |
4WHY
 
 | |
4UDQ
 
 | | Crystal structure of 5-hydroxymethylfurfural oxidase (HMFO) in the reduced state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLUCOSE-METHANOL-CHOLINE OXIDOREDUCTASE | | Authors: | Dijkman, W, Binda, C, Fraaije, M, Mattevi, A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Enzyme Tailoring of 5-Hydroxymethylfurfural Oxidase
Acs Catalysis, 5, 2015
|
|
