6T3L
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome in dark state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
6I29
 
 | | X-ray structure of the p53-MDM2 inhibitor NMI801 bound to HDM2 at 2.1A resolution | | Descriptor: | 6-chloranyl-3-[3-[(1~{S})-1-(4-chlorophenyl)ethyl]-5-phenyl-imidazol-4-yl]-~{N}-[2-[4-(2-oxidanylidene-1,3-oxazinan-3-yl)piperidin-1-yl]pyridin-3-yl]-1~{H}-indole-2-carboxamide, Human E3 Ubiquitin-Protein Ligase MDM2 | | Authors: | Kallen, J. | | Deposit date: | 2018-11-01 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | p53 dynamics vary between tissues and are linked with radiation sensitivity
To be published
|
|
6T3U
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome 1ps after photoexcitation | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
5JAQ
 
 | | Yersinia pestis FabV variant T276C | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
6VFX
 
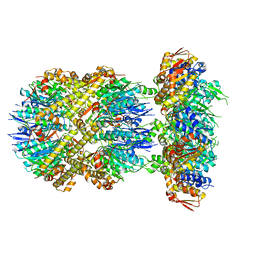 | | ClpXP from Neisseria meningitidis - Conformation B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
4YGY
 
 | |
5LXZ
 
 | |
6I51
 
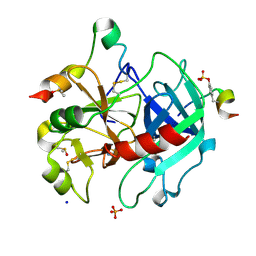 | | Thrombin in complex with fragment J02 | | Descriptor: | 1H-isoindol-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Abazi, N, Heine, A, Klebe, G. | | Deposit date: | 2018-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment screening of Thrombin using a 96 member fragment library
To Be Published
|
|
7YDG
 
 | | Crystal structure of human SARS2 catalytic domain with a disease related mutation | | Descriptor: | Serine--tRNA ligase, mitochondrial | | Authors: | Wu, S, Li, P, Zhou, X.L, Fang, P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
7YDF
 
 | | Crystal structure of human SARS2 catalytic domain | | Descriptor: | Serine--tRNA ligase, mitochondrial | | Authors: | Wu, S, Li, P, Zhou, X.L, Fang, P. | | Deposit date: | 2022-07-04 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective degradation of tRNASer(AGY) is the primary driver for mitochondrial seryl-tRNA synthetase-related disease.
Nucleic Acids Res., 50, 2022
|
|
7Y4K
 
 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-ornitine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-5-(phenylmethoxycarbonylamino)pentanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
7Y4M
 
 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-lysine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-6-(phenylmethoxycarbonylamino)hexanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
6IM4
 
 | |
3AU8
 
 | | Crystal structure of the ternary complex of an isomerase | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Umeda, T, Tanaka, N, Kusakabe, Y, Nakanishi, M, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2011-02-01 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular basis of fosmidomycin's action on the human malaria parasite Plasmodium falciparum
Sci Rep, 1, 2011
|
|
6VFS
 
 | | ClpXP from Neisseria meningitidis - Conformation A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ripstein, Z.A, Vahidi, S, Houry, W.A, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-06 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A processive rotary mechanism couples substrate unfolding and proteolysis in the ClpXP degradation machinery.
Elife, 9, 2020
|
|
7VF6
 
 | | The crystal structure of PurZ0 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Tong, Y, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Alternative Z-genome biosynthesis pathway shows evolutionary progression from Archaea to phage.
Nat Microbiol, 8, 2023
|
|
3M4P
 
 | |
7Z8O
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, GLYCEROL, Spike protein S1, ... | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-03-18 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
3IDQ
 
 | | Crystal structure of S. cerevisiae Get3 at 3.7 Angstrom resolution | | Descriptor: | ATPase GET3, NICKEL (II) ION, ZINC ION | | Authors: | Suloway, C.J.M, Chartron, J.W, Zaslaver, M, Clemons Jr, W.M. | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Model for eukaryotic tail-anchored protein binding based on the structure of Get3
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1RMY
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase ternary complex with deoxycytosine and phosphate bound to the catalytic metal | | Descriptor: | 2'-DEOXYCYTIDINE, Class B acid phosphatase, MAGNESIUM ION, ... | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
1RBB
 
 | |
6URS
 
 | | Sleeping Beauty transposase PAI subdomain mutant - H19Y | | Descriptor: | Sleeping Beauty transposase PAI subdomain | | Authors: | Nesmelova, I.V, Leighton, G.O, Yan, C, Lustig, J, Corona, R.I, Guo, J.T, Ivics, Z. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | H19Y mutation in the primary DNA-recognition subdomain of the Sleeping Beauty transposase improves structural stability, transposon DNA-binding and transposition
To Be Published
|
|
3M4Q
 
 | |
1IKA
 
 | |
1IXP
 
 | | Enzyme-phosphate Complex of Pyridoxine 5'-Phosphate synthase | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-Phosphate synthase | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
