1DEE
 
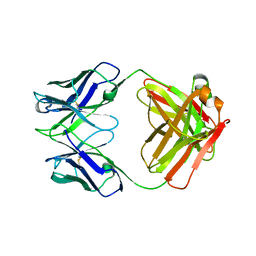 | | Structure of S. aureus protein A bound to a human IgM Fab | | Descriptor: | IGM RF 2A2, IMMUNOGLOBULIN G BINDING PROTEIN A | | Authors: | Graille, M, Stura, E.A, Corper, A.L, Sutton, B.J, Taussig, M.J, Charbonnier, J.B, Silverman, G.J. | | Deposit date: | 1999-11-15 | | Release date: | 2000-05-24 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EG9
 
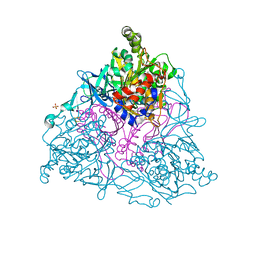 | | NAPHTHALENE 1,2-DIOXYGENASE WITH INDOLE BOUND IN THE ACTIVE SITE. | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, INDOLE, ... | | Authors: | Carredano, E, Karlsson, A, Kauppi, B, Choudhury, D, Parales, R.E, Parales, J.V, Lee, K, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2000-02-15 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding.
J.Mol.Biol., 296, 2000
|
|
1EF8
 
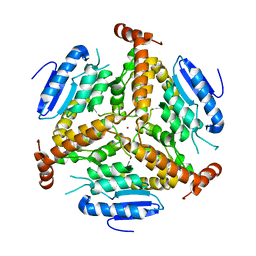 | | CRYSTAL STRUCTURE OF METHYLMALONYL COA DECARBOXYLASE | | Descriptor: | METHYLMALONYL COA DECARBOXYLASE, NICKEL (II) ION | | Authors: | Benning, M.M, Haller, T, Gerlt, J.A, Holden, H.M. | | Deposit date: | 2000-02-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1EZ0
 
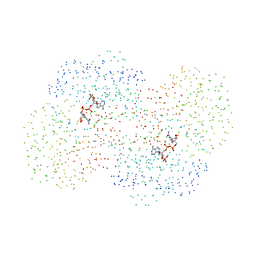 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | Descriptor: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
1EYY
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | Descriptor: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
1EZK
 
 | | Crystal structure of recombinant tryparedoxin I | | Descriptor: | TRYPAREDOXIN I | | Authors: | Hofmann, B, Guerrero, S.A, Kalisz, H.M, Menge, U, Nogoceke, E, Montemartini, M, Singh, M, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
1D0U
 
 | |
1D2D
 
 | | Hamster EprS second repeated element. NMR, 5 structures | | Descriptor: | TRNA SYNTHETASE | | Authors: | Cahuzac, B, Berthonneau, E, Birlirakis, N, Guittet, E, Mirande, M. | | Deposit date: | 1999-09-23 | | Release date: | 2000-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases.
EMBO J., 19, 2000
|
|
1D0T
 
 | |
1EF9
 
 | | THE CRYSTAL STRUCTURE OF METHYLMALONYL COA DECARBOXYLASE COMPLEXED WITH 2S-CARBOXYPROPYL COA | | Descriptor: | 2-CARBOXYPROPYL-COENZYME A, METHYLMALONYL COA DECARBOXYLASE | | Authors: | Benning, M.M, Haller, T, Gerlt, J.A, Holden, H.M. | | Deposit date: | 2000-02-07 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli.
Biochemistry, 39, 2000
|
|
1QPD
 
 | | STRUCTURAL ANALYSIS OF THE LYMPHOCYTE-SPECIFIC KINASE LCK IN COMPLEX WITH NON-SELECTIVE AND SRC FAMILY SELECTIVE KINASE INHIBITORS | | Descriptor: | LCK KINASE, STAUROSPORINE, SULFATE ION | | Authors: | Zhu, X, Kim, J.L, Rose, P.E, Stover, D.R, Toledo, L.M, Zhao, H, Morgenstern, K.A. | | Deposit date: | 1999-05-24 | | Release date: | 2000-05-24 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors.
Structure Fold.Des., 7, 1999
|
|
1CN0
 
 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Cheng, Y, Wang, S, Sutherland, C, Sinha, N, Kang, C. | | Deposit date: | 1999-05-24 | | Release date: | 2000-05-24 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QHM
 
 | | ESCHERICHIA COLI PYRUVATE FORMATE LYASE LARGE DOMAIN | | Descriptor: | PYRUVATE FORMATE-LYASE | | Authors: | Leppanen, V.-M, Merckel, M.C, Ollis, D.L, Wong, K.K, Kozarich, J.W, Goldman, A. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyruvate formate lyase is structurally homologous to type I ribonucleotide reductase.
Structure Fold.Des., 7, 1999
|
|
1QPC
 
 | | STRUCTURAL ANALYSIS OF THE LYMPHOCYTE-SPECIFIC KINASE LCK IN COMPLEX WITH NON-SELECTIVE AND SRC FAMILY SELECTIVE KINASE INHIBITORS | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Zhu, X, Kim, J.L, Rose, P.E, Stover, D.R, Toledo, L.M, Zhao, H, Morgenstern, K.A. | | Deposit date: | 1999-05-21 | | Release date: | 2000-05-24 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of the lymphocyte-specific kinase Lck in complex with non-selective and Src family selective kinase inhibitors.
Structure Fold.Des., 7, 1999
|
|
1QQ3
 
 | | THE SOLUTION STRUCTURE OF THE HEME BINDING VARIANT ARG98CYS OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, HEME B/C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Ciofi-Baffoni, S, Barker, P.D, Woodyear, T. | | Deposit date: | 1999-06-10 | | Release date: | 2000-05-24 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structural consequences of b- to c-type heme conversion in oxidized Escherichia coli cytochrome b562.
Biochemistry, 39, 2000
|
|
1QME
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1QMF
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
1DZH
 
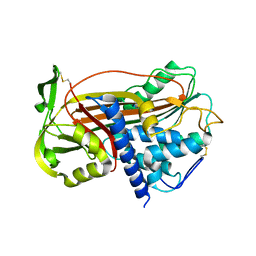 | | P14-FLUORESCEIN-N135Q-S380C-ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | Mccoy, A.J, Huntington, J.A, Carrell, R.W. | | Deposit date: | 2000-02-28 | | Release date: | 2000-05-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Conformational Activation of Antithrombin. A 2. 85-A Structure of a Fluorescein Derivative Reveals an Electrostatic Link between the Hinge and Heparin Binding Regions.
J.Biol.Chem., 275, 2000
|
|
1DZG
 
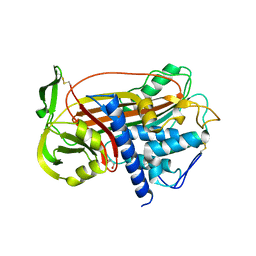 | | N135Q-S380C-ANTITHROMBIN-III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTITHROMBIN-III, ... | | Authors: | McCoy, A.J, Huntington, J.A, Carrell, R.W. | | Deposit date: | 2000-02-28 | | Release date: | 2000-05-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Conformational Activation of Antithrombin. A 2. 85-A Structure of a Fluorescein Derivative Reveals an Electrostatic Link between the Hinge and Heparin Binding Regions.
J.Biol.Chem., 275, 2000
|
|
1QHT
 
 | |
1QOW
 
 | |
1EQ6
 
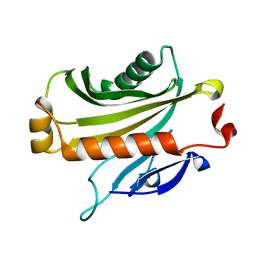 | |
1EFO
 
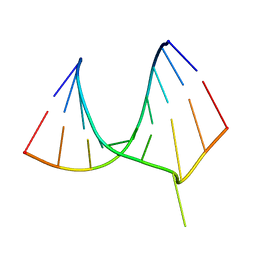 | | CRYSTAL STRUCTURE OF AN ADENINE BULGE IN THE RNA CHAIN OF A DNA/RNA HYBRID, D(CTCCTCTTC)/R(GAAGAGAGAG) | | Descriptor: | DNA (5'-D(*CP*TP*CP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3') | | Authors: | Sudarsanakumar, C, Xiong, Y, Sundaralingam, M. | | Deposit date: | 2000-02-09 | | Release date: | 2000-05-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an adenine bulge in the RNA chain of a DNA.RNA hybrid, d(CTCCTCTTC).r(gaagagagag).
J.Mol.Biol., 299, 2000
|
|
419D
 
 | |
1E2R
 
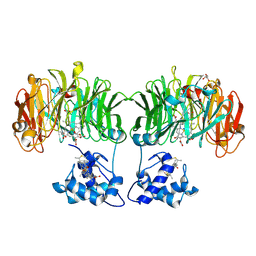 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED AND CYANIDE BOUND | | Descriptor: | CYANIDE ION, GLYCEROL, HEME C, ... | | Authors: | Fulop, V. | | Deposit date: | 2000-05-24 | | Release date: | 2000-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-Ray Crystallographic Study of Cyanide Binding Provides Insights Into the Structure-Function Relationship for Cytochrome Cd1 Nitrite Reductase from Paracoccus Pantotrophus.
J.Biol.Chem., 275, 2000
|
|
