6VGQ
 
 | | ClpP1P2 complex from M. tuberculosis with GLF-CMK bound to ClpP1 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, ATP-dependent Clp protease proteolytic subunit 1, Z-Gly-leu-phe-CH2Cl | | Authors: | Ripstein, Z.A, Vahidi, S, Rubinstein, J.L, Kay, L.E. | | Deposit date: | 2020-01-08 | | Release date: | 2020-03-18 | | Last modified: | 2020-04-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An allosteric switch regulatesMycobacterium tuberculosisClpP1P2 protease function as established by cryo-EM and methyl-TROSY NMR.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8SE9
 
 | | Cryo-EM structure of a double loaded human UBA7-UBE2L6-ISG15 thioester mimetic complex (Form 2) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin-like modifier-activating enzyme 7, Ubiquitin-like protein ISG15, ... | | Authors: | Afsar, M, Jia, L, Ruben, E.A, Olsen, S.K. | | Deposit date: | 2023-04-08 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Uba7 reveal the molecular basis for ISG15 activation and E1-E2 thioester transfer.
Nat Commun, 14, 2023
|
|
6UPP
 
 | | Radiation Damage Test of PixJ Pb state crystals | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Burgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4PYY
 
 | | Crystal structure of human carbonic anhydrase isozyme II with inhibitor | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-03-28 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and characterization of novel selective inhibitors of carbonic anhydrase IX.
J.Med.Chem., 57, 2014
|
|
7TBM
 
 | | Composite structure of the dilated human nuclear pore complex (NPC) generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | Descriptor: | DDX19, NUP107 CTD, NUP107 NTD, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
6Y55
 
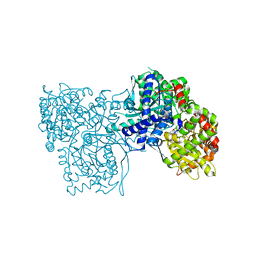 | | The crystal structure of glycogen phosphorylase in complex with 43 | | Descriptor: | 2-(3-methylphenyl)-5,7-bis(oxidanyl)chromen-4-one, Glycogen phosphorylase, muscle form | | Authors: | Kyriakis, E, Koulas, S.M, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2020-02-24 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Synthetic flavonoid derivatives targeting the glycogen phosphorylase inhibitor site: QM/MM-PBSA motivated synthesis of substituted 5,7-dihydroxyflavones, crystallography, in vitro kinetics and ex-vivo cellular experiments reveal novel potent inhibitors.
Bioorg.Chem., 102, 2020
|
|
6FA2
 
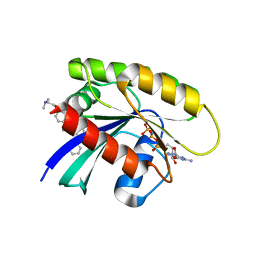 | | Antibody derived (Abd-5) small molecule binding to KRAS. | | Descriptor: | 4-[2-(dimethylamino)ethoxy]-~{N}-[[(3~{R})-5-(6-methoxypyridin-2-yl)-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]benzamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Quevedo, C.E, Cruz-Migoni, A, Ehebauer, M.T, Carr, S.B, Phillips, S.V.E, Rabbitts, T.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
6PB0
 
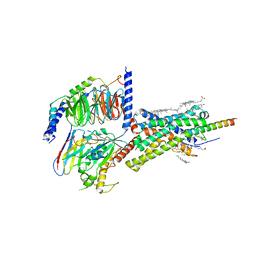 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
6MJ5
 
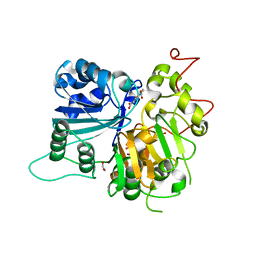 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ519 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-nitroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
5AQR
 
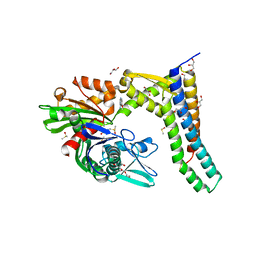 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-methoxyquinazolin-4-amine, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
7TA2
 
 | | Crystal structure of the human sperm-expressed surface protein SPACA6 | | Descriptor: | BROMIDE ION, Sperm acrosome membrane-associated protein 6 | | Authors: | Vance, T.D.R, Yip, P, Jimenez, E, Uson, I, Lee, J.E. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins.
Commun Biol, 5, 2022
|
|
6PBF
 
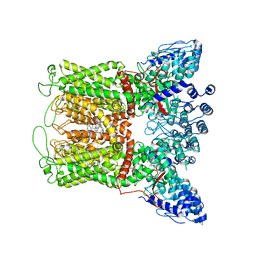 | | ZINC9155420-bound TRPV5 in nanodiscs | | Descriptor: | (1S)-1-(4-oxo-5-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-2-yl)ethyl [(2S)-3-oxo-3,4-dihydro-2H-1,4-benzothiazin-2-yl]acetate, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Rosario, J.S.D, Kapoor, A, Yazici, A.T, Fluck, E.C, Filizola, M, Rohacs, T, Moiseenkova-Bell, V.Y. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure-based characterization of novel TRPV5 inhibitors.
Elife, 8, 2019
|
|
1OUZ
 
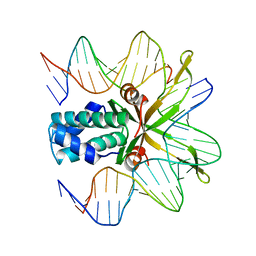 | | Crystal structure of a mutant IHF (BetaE44A) complexed with a variant H' Site (T44A) | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*AP*GP*CP*AP*CP*C)-3', 5'-D(*GP*GP*CP*CP*AP*AP*AP*AP*AP*AP*GP*CP*AP*TP*T)-3', Integration Host Factor Alpha-subunit, ... | | Authors: | Lynch, T.W, Read, E.K, Mattis, A.N, Gardner, J.F, Rice, P.A. | | Deposit date: | 2003-03-25 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Integration Host Factor: putting a twist on protein-DNA recognition
J.Mol.Biol., 330, 2003
|
|
6FF7
 
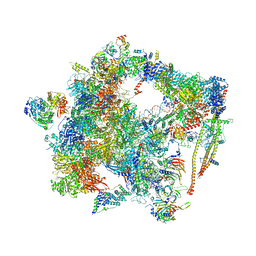 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, BUD13 homolog, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
6F5H
 
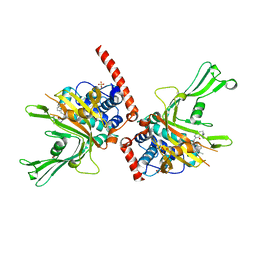 | | Crystal structure of USP7 in complex with a 4-hydroxypiperidine based inhibitor | | Descriptor: | 3-[[4-oxidanyl-1-[(3~{R})-3-phenylbutanoyl]piperidin-4-yl]methyl]-6-(2-pyrrolidin-1-ylethylamino)pyrimidin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Harrison, T, Gavory, G, O'Dowd, C, Helm, M, Flasz, J, Dossang, A, Hughes, C, Cassidy, E, McClelland, K, Odrzywol, E, Page, N, Barker, O, Miel, H, Feutron-Burton, S, Rountree, J.S.S. | | Deposit date: | 2017-12-01 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Identification and Structure-Guided Development of Pyrimidinone Based USP7 Inhibitors.
ACS Med Chem Lett, 9, 2018
|
|
6PBE
 
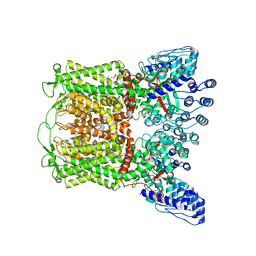 | | ZINC17988990-bound TRPV5 in nanodiscs | | Descriptor: | (4-oxo-5-phenyl-3,4-dihydrothieno[2,3-d]pyrimidin-2-yl)methyl 3-(3-oxo-2,3-dihydro-4H-1,4-benzoxazin-4-yl)propanoate, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Rosario, J.S.D, Kapoor, A, Yazici, A.T, Fluck, E.C, Filizola, M, Rohacs, T, Moiseenkova-Bell, V.Y. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structure-based characterization of novel TRPV5 inhibitors.
Elife, 8, 2019
|
|
6MHB
 
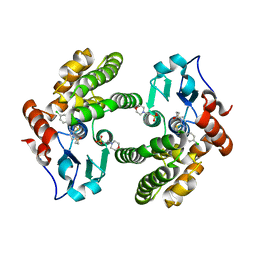 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 18 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-[4-(4-chlorophenyl)-1,3-thiazol-2-yl]propanamide | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6UXR
 
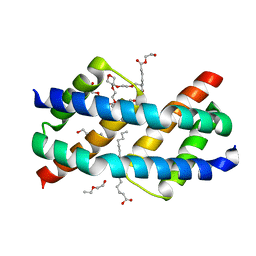 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with LysoPC | | Descriptor: | Bcl-2 homologous antagonist/killer, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8ADU
 
 | | Lipidic alpha-synuclein fibril - polymorph L1A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADS
 
 | | Lipidic alpha-synuclein fibril - polymorph L2B | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
7OFW
 
 | | Nontypeable Haemophillus influenzae SapA in complex with heme | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
5FT8
 
 | | Crystal structure of the complex between the cysteine desulfurase CsdA and the sulfur-acceptor CsdE in the persulfurated state at 2.50 Angstroem resolution | | Descriptor: | Cysteine desulfurase CsdA, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
6Y5C
 
 | | The crystal structure of glycogen phosphorylase in complex with 52 | | Descriptor: | 2-(4-methylphenyl)-5,7-bis(oxidanyl)chromen-4-one, Glycogen phosphorylase, muscle form | | Authors: | Kyriakis, E, Koulas, S.M, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2020-02-25 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthetic flavonoid derivatives targeting the glycogen phosphorylase inhibitor site: QM/MM-PBSA motivated synthesis of substituted 5,7-dihydroxyflavones, crystallography, in vitro kinetics and ex-vivo cellular experiments reveal novel potent inhibitors.
Bioorg.Chem., 102, 2020
|
|
8DK6
 
 | | Structure of hepatitis C virus envelope N-terminal truncated glycoprotein 2 (E2) (residues 456-713) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, 2A12 Fab light chain, ... | | Authors: | Kumar, A, Rohe, T, Elrod, E.J, Khan, A.G, Dearborn, A.D, Kissinger, R, Grakoui, A, Marcotrigiano, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regions of hepatitis C virus E2 required for membrane association.
Nat Commun, 14, 2023
|
|
6PA7
 
 | | The cryo-EM structure of the human DNMT3A2-DNMT3B3 complex bound to nucleosome. | | Descriptor: | CHLORIDE ION, DNA (167-MER), DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Xu, T.H, Liu, M, Zhou, X.E, Liang, G, Zhao, G, Xu, H.E, Melcher, K, Jones, P.A. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B.
Nature, 586, 2020
|
|
