4CI3
 
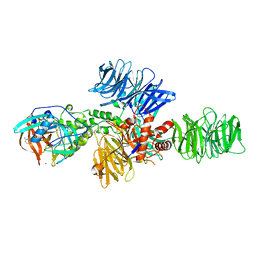 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
3OFJ
 
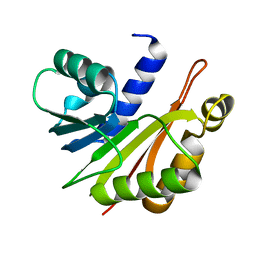 | | Crystal structure of N-methyltransferase NodS from Bradyrhizobium japonicum WM9 | | Descriptor: | Nodulation protein S | | Authors: | Cakici, O, Sikorski, M, Stepkowski, T, Bujacz, G, Jaskolski, M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of NodS N-Methyltransferase from Bradyrhizobium japonicum in Ligand-Free Form and as SAH Complex.
J.Mol.Biol., 404, 2010
|
|
4FQK
 
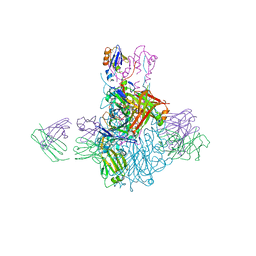 | | Influenza B/Brisbane/60/2008 hemagglutinin Fab CR8059 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody CR8059 Heavy Chain, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5.65 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQJ
 
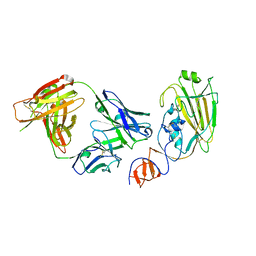 | | Influenza B/Florida/4/2006 hemagglutinin Fab CR8071 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, antibody CR8071 heavy chain, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQY
 
 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H3 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.253 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQI
 
 | |
3PFF
 
 | |
4FS4
 
 | | Structure of BACE Bound to (S)-4-(3'-methoxy-[1,1'-biphenyl]-3-yl)-1,4-dimethyl-6-oxotetrahydropyrimidin-2(1H)-iminium | | Descriptor: | (6S)-2-amino-6-(3'-methoxybiphenyl-3-yl)-3,6-dimethyl-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Stamford, A. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-10 | | Last modified: | 2014-07-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
4GAV
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with quinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase, UBIQUINONE-2 | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8FFX
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 19980 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Reverse transcriptase/ribonuclease H, ... | | Authors: | Rumrill, S.R, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Targeting HIV-1 Reverse Transcriptase Using a Fragment-Based Approach.
Molecules, 28, 2023
|
|
5OVR
 
 | | X-Ray Characterization of Striatal-Enriched Protein Tyrosine Phosphatase Inhibitors | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 5, [(~{S})-[4-[3-[(~{R})-(3,4-dichlorophenyl)-oxidanyl-methyl]phenyl]phenyl]-oxidanyl-methyl]phosphonic acid | | Authors: | Kack, H, Wissler, L. | | Deposit date: | 2017-08-29 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Characterization and Structure-Based Optimization of Striatal-Enriched Protein Tyrosine Phosphatase Inhibitors.
J. Med. Chem., 60, 2017
|
|
6OF5
 
 | | The crystal structure of dodecyloxy(naphthalen-1-yl)methylphosphonic acid in complex with red kidney bean purple acid phosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, Hussein, W.M, McGeary, R.P. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis, evaluation and structural investigations of potent purple acid phosphatase inhibitors as drug leads for osteoporosis.
Eur.J.Med.Chem., 182, 2019
|
|
5OVX
 
 | | X-Ray Characterization of Striatal-Enriched Protein Tyrosine Phosphatase Inhibitors | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 5, [(~{S})-[4-[3-[(~{S})-(3,4-dichlorophenyl)-oxidanyl-methyl]phenyl]phenyl]-oxidanyl-methyl]phosphonic acid | | Authors: | Kack, H, Wissler, L. | | Deposit date: | 2017-08-30 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Characterization and Structure-Based Optimization of Striatal-Enriched Protein Tyrosine Phosphatase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5OW1
 
 | |
6OWE
 
 | | Enoyl-CoA carboxylases/reductases in complex with ethylmalonyl CoA | | Descriptor: | 5'-O-[(S)-{[(S)-[(3R)-4-({(1E)-3-[(2-{[(2S)-2-carboxybutanoyl]sulfanyl}ethyl)amino]-3-oxoprop-1-en-1-yl}amino)-3-hydroxy-2,2-dimethyl-4-oxobutoxy](hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]adenosine 3'-(dihydrogen phosphate), Crotonyl-CoA carboxylase/reductase, IMIDAZOLE, ... | | Authors: | DeMirci, H. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Four amino acids define the CO2binding pocket of enoyl-CoA carboxylases/reductases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4HW1
 
 | | Multiple Crystal structures of an all-AT DNA dodecamer stabilized by weak interactions. | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3'), MAGNESIUM ION | | Authors: | Acosta-Reyes, F, Subirana, J.A, Pous, J, Condom, N, Malinina, L, Campos, J.L. | | Deposit date: | 2012-11-07 | | Release date: | 2013-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
6OB1
 
 | | Structure of WHB in complex with Ubiquitin Variant | | Descriptor: | Anaphase-promoting complex subunit 2, Ubiquitin | | Authors: | Edmond, R.W, Grace, C.R. | | Deposit date: | 2019-03-19 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein engineering of a ubiquitin-variant inhibitor of APC/C identifies a cryptic K48 ubiquitin chain binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5PTI
 
 | |
6PCA
 
 | | Crystal structure of beta-ketoadipyl-CoA thiolase | | Descriptor: | ACETATE ION, Beta-ketoadipyl-CoA thiolase, CHLORIDE ION, ... | | Authors: | Sukritee, B, Panjikar, S. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for differentiation between two classes of thiolase: Degradative vs biosynthetic thiolase.
J Struct Biol X, 4, 2020
|
|
8G5C
 
 | |
8G92
 
 | | Structure of inhibitor 16d-bound SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | Authors: | Chen, H, Li, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
8G5D
 
 | |
6PFN
 
 | | Succinyl-CoA synthase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COENZYME A, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Succinyl-CoA synthase from Francisella tularensis
to be published
|
|
6PCB
 
 | |
4J4R
 
 | | Hexameric SFTSVN | | Descriptor: | Nucleocapsid protein | | Authors: | Jiao, L, Ouyang, S, Liang, M, Niu, F, Shaw, N, Wu, W, Ding, W, Jin, C, Zhu, Y, Zhang, F, Wang, T, Li, C, Zuo, X, Luan, C.H, Li, D, Liu, Z.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of severe Fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential
J.Virol., 87, 2013
|
|
