4GET
 
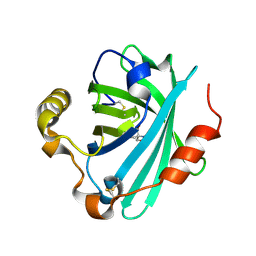 | | Crystal structure of biogenic amine binding protein from Rhodnius prolixus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Biogenic amine-binding protein | | Authors: | Andersen, J.F, Chang, B.W, Xu, X, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2ANJ
 
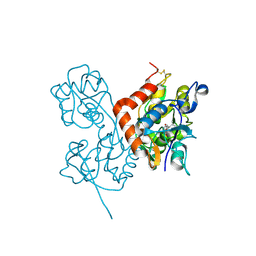 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
1UX8
 
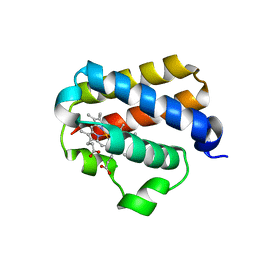 | | X-ray structure of truncated oxygen-avid haemoglobin from Bacillus subtilis | | Descriptor: | CHLORIDE ION, CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ilari, A, Giangiacomo, L, Boffi, A, Chiancone, E. | | Deposit date: | 2004-02-20 | | Release date: | 2004-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Truncated Oxygen-Avid Hemoglobin from Bacillus Subtilis: X-Ray Structure and Ligand Binding Properties
J.Biol.Chem., 280, 2005
|
|
6ELX
 
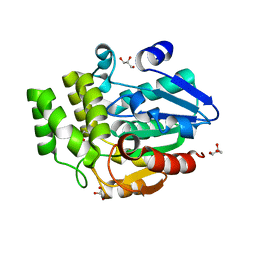 | | Oryza sativa DWARF14 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Andersson, I, Carlsson, G.H, Hasse, D. | | Deposit date: | 2017-09-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The elusive ligand complexes of the DWARF14 strigolactone receptor.
J. Exp. Bot., 69, 2018
|
|
5H9Q
 
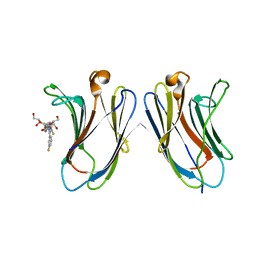 | | Crystal Structure of Human Galectin-7 in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-7 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
4DE1
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 18 | | Descriptor: | Beta-lactamase, DIMETHYL SULFOXIDE, N-[3-(1H-tetrazol-5-yl)phenyl]-2H-indazole-5-carboxamide | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
6QIQ
 
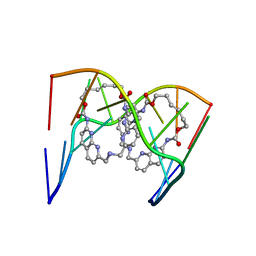 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL3a compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*G)-D(P*(CSL))-R(P*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Micura, R, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIT
 
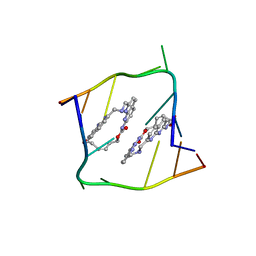 | | Crystal structure of CAG repeats with synthetic CMBL3b compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
5H9R
 
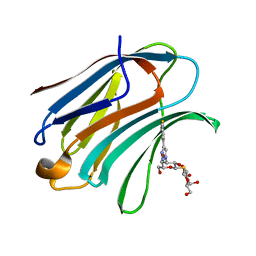 | | Crystal Structure of Human Galectin-3 CRD in Complex with TAZTDG | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
6HCD
 
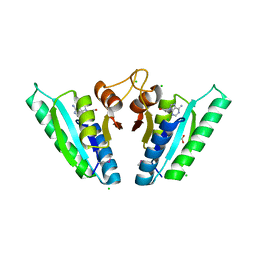 | | Structure of universal stress protein from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | Authors: | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-08-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
6QIR
 
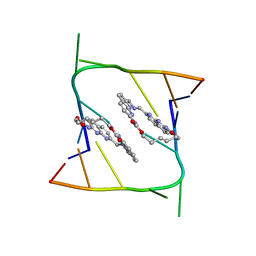 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model I) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
5H9P
 
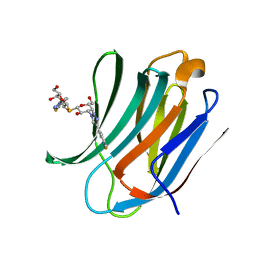 | | Crystal Structure of Human Galectin-3 CRD in Complex with TD139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-3 | | Authors: | Hsieh, T.J, Lin, H.Y, Lin, C.H. | | Deposit date: | 2015-12-29 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors
Sci Rep, 6, 2016
|
|
3M6Q
 
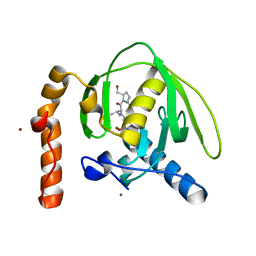 | |
3MFA
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
4BSK
 
 | | Crystal structure of VEGF-C in complex with VEGFR-3 domains D1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, VASCULAR ENDOTHELIAL GROWTH FACTOR C, ... | | Authors: | Leppanen, V.M, Tvorogov, D, Kisko, K, Prota, A.E, Jeltsch, M, Anisimov, A, Markovic-Mueller, S, Stuttfeld, E, Goldie, K.N, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.201 Å) | | Cite: | Structural and Mechanistic Insights Into Vegfr-3 Ligand Binding and Activation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3M6P
 
 | |
4DE2
 
 | | CTX-M-9 class A beta-lactamase complexed with compound 12 | | Descriptor: | 3-[(dimethylamino)methyl]-N-[3-(1H-tetrazol-5-yl)phenyl]benzamide, Beta-lactamase, DIMETHYL SULFOXIDE | | Authors: | Nichols, D.A, Chen, Y. | | Deposit date: | 2012-01-19 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Design of Potent and Ligand-Efficient Inhibitors of CTX-M Class A Beta-Lactamase
J.Med.Chem., 55, 2012
|
|
3MF6
 
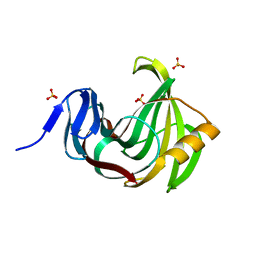 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MFC
 
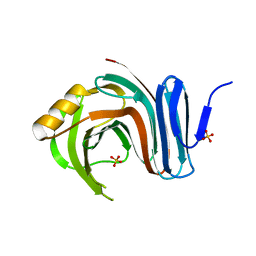 | | Computationally designed end0-1,4-beta,xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3GYR
 
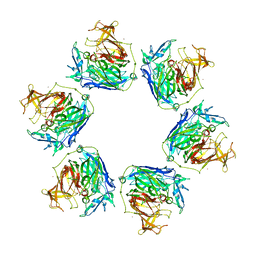 | | Structure of Phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, GLYCEROL, ... | | Authors: | Smith, A.W, Camara-Artigas, A, Wang, M, Francisco, W.A, Allen, J.P. | | Deposit date: | 2009-04-05 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Phenoxazinone Synthase from Streptomyces Antibioticus Reveals a New Type 2 Copper Center.
Biochemistry, 45, 2006
|
|
3VHV
 
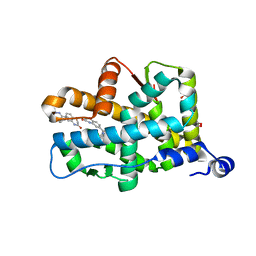 | | Mineralocorticoid receptor ligand-binding domain with non-steroidal antagonist | | Descriptor: | 1,2-ETHANEDIOL, 6-[(1E)-2-phenyl-N-(3-sulfanyl-4H-1,2,4-triazol-4-yl)ethanimidoyl]-2H-1,4-benzoxazin-3(4H)-one, 6-[(7S)-7-phenyl-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazin-6-yl]-2H-1,4-benzoxazin-3(4H)-one, ... | | Authors: | Sogabe, S, Habuka, N. | | Deposit date: | 2011-09-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of Benzoxazin-3-one Derivatives as Novel, Potent, and Selective Nonsteroidal Mineralocorticoid Receptor Antagonists
J.Med.Chem., 54, 2011
|
|
4KE7
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-myristoyl glycerol analogue | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase, dodecyl hydrogen (S)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KEA
 
 | | Crystal structure of D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in space group P212121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
3ISS
 
 | | Crystal structure of enolpyruvyl-UDP-GlcNAc synthase (MurA):UDP-N-acetylmuramic acid:phosphite from Escherichia coli | | Descriptor: | PHOSPHITE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Jackson, S.G, Zhang, F, Chindemi, P, Junop, M.S, Berti, P.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence of Kinetic Control of Ligand Binding and Staged Product Release in MurA (Enolpyruvyl UDP-GlcNAc Synthase)-Catalyzed Reactions .
Biochemistry, 48, 2009
|
|
2MM3
 
 | | Solution NMR structure of the ternary complex of human ileal bile acid-binding protein with glycocholate and glycochenodeoxycholate | | Descriptor: | GLYCOCHENODEOXYCHOLIC ACID, GLYCOCHOLIC ACID, Gastrotropin | | Authors: | Horvath, G, Egyed, O, Bencsura, A, Simon, A, Tochtrop, G.P, DeKoster, G.T, Covey, D.F, Cistola, D.P, Toke, O. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of ligand binding in the ternary complex of human ileal bile acid binding protein with glycocholate and glycochenodeoxycholate obtained from solution NMR
FEBS J., 283, 2016
|
|
