7ZN4
 
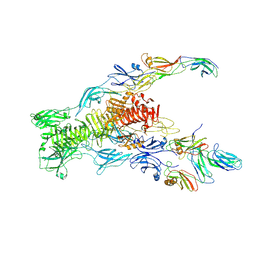 | | Tail tip of siphophage T5 : bent fibre after interaction with its bacterial receptor FhuA | | Descriptor: | Probable baseplate hub protein, Probable central straight fiber | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-20 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZQB
 
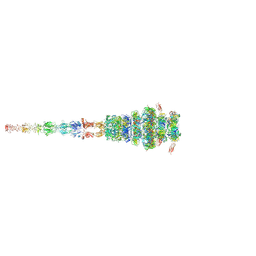 | | Tail tip of siphophage T5 : full structure | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZN2
 
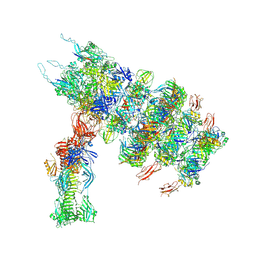 | | Tail tip of siphophage T5 : full complex after interaction with its bacterial receptor FhuA | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-20 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7UVW
 
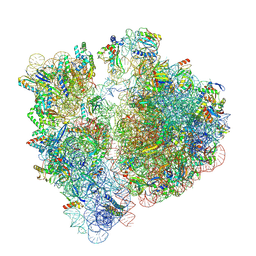 | | A. baumannii ribosome: 70S with E-site tRNA | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-19 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome.
Plos Biol., 21, 2023
|
|
9F3Q
 
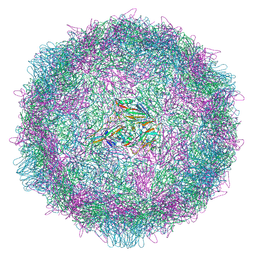 | | Poliovirus type 1 (strain Mahoney) stabilised virus-like particle (PV1 SC6b) in complex with GPP3 and GSH. | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein VP0, Capsid protein VP1, ... | | Authors: | Bahar, M.W, Nasta, V, Sherry, L, Stonehouse, N.J, Rowlands, D.J, Fry, E.E, Stuart, D.I. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Recombinant expression systems for production of stabilised virus-like particles as next-generation polio vaccines.
Nat Commun, 16, 2025
|
|
9B0T
 
 | | Cryo-EM structure of E227Q variant of uMtCK1 in complex with transition state analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-12 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
9B14
 
 | | Cryo-EM structure of human uMtCK1 in complex with transition state analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-13 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
9B0U
 
 | | Cryo-EM structure of E227Q variant of uMtCK1 incubated with ADP and phosphocreatine at pH 8.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-12 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
5JB3
 
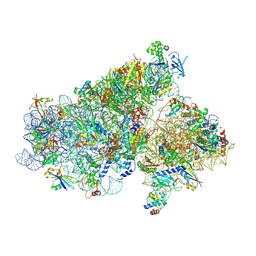 | | Cryo-EM structure of a full archaeal ribosomal translation initiation complex in the P-REMOTE conformation | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Schmitt, E, Mechulam, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Cryo-EM study of start codon selection during archaeal translation initiation.
Nat Commun, 7, 2016
|
|
2XO4
 
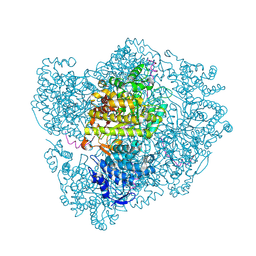 | | RIBONUCLEOTIDE REDUCTASE Y730NH2Y MODIFIED R1 SUBUNIT OF E. COLI | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT ALPHA, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 SUBUNIT BETA | | Authors: | Minnihan, E.C, Seyedsayamdost, M.R, Uhlin, U, Stubbe, J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinetics of Radical Intermediate Formation and Deoxynucleotide Production in 3-Aminotyrosine- Substituted Escherichia Coli Ribonucleotide Reductases.
J.Am.Chem.Soc., 133, 2011
|
|
5UQ7
 
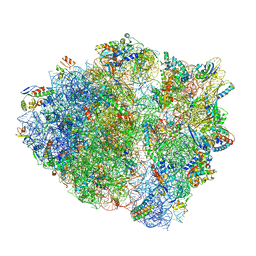 | | 70S ribosome complex with dnaX mRNA stemloop and E-site tRNA ("in" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
2KNY
 
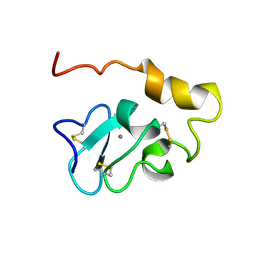 | |
3DW5
 
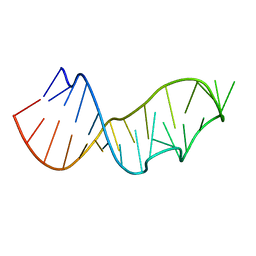 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23S rRNA, U2656-OCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
5KDI
 
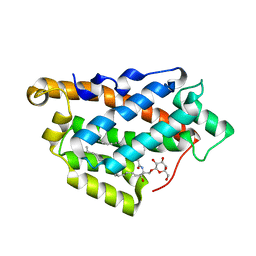 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
6U2M
 
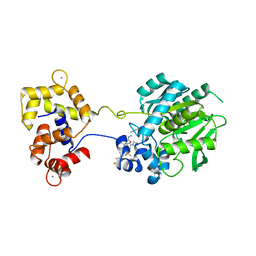 | | Crystal structure of a HaloTag-based calcium indicator, HaloCaMP V2, bound to JF635 | | Descriptor: | (1E,3S)-1-{10-[2-carboxy-5-({2-[2-(hexyloxy)ethoxy]ethyl}carbamoyl)phenyl]-7-(3-fluoroazetidin-1-yl)-5,5-dimethyldibenz o[b,e]silin-3(5H)-ylidene}-3-fluoroazetidin-1-ium, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Deo, C, Schreiter, E.R. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The HaloTag as a general scaffold for far-red tunable chemigenetic indicators.
Nat.Chem.Biol., 17, 2021
|
|
5KPV
 
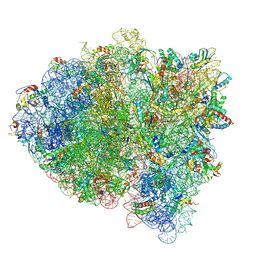 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
5KPW
 
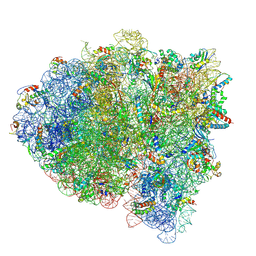 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
6DL7
 
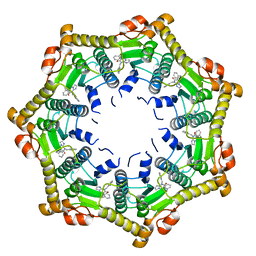 | | Human mitochondrial ClpP in complex with ONC201 (TIC10) | | Descriptor: | 7-benzyl-4-[(2-methylphenyl)methyl]-6,7,8,9-tetrahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(4H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Halgas, O, Zarabi, S.F, Schimmer, A, Pai, E.F. | | Deposit date: | 2018-05-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality.
Cancer Cell, 35, 2019
|
|
5UQ8
 
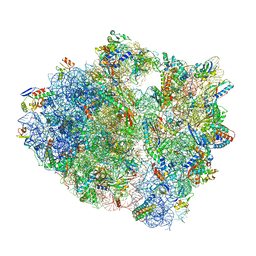 | | 70S ribosome complex with dnaX mRNA stem-loop and E-site tRNA ("out" conformation) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, Y, Hong, S, Skiniotis, G, Dunham, C.M. | | Deposit date: | 2017-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Alternative Mode of E-Site tRNA Binding in the Presence of a Downstream mRNA Stem Loop at the Entrance Channel.
Structure, 26, 2018
|
|
5F6C
 
 | |
5LQR
 
 | | Crystal Structure of COMT in complex with N-[(E)-3-[(2R,3S,4R,5R)-5-(6-ethylpurin-9-yl)-3,4-dihydroxyoxolan-2-yl]prop-2-enyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of COMT in complex with N-[(E)-3-[(2R,3S,4R,5R)-5-(6-ethylpurin-9-yl)-3,4-dihydroxyoxolan-2-yl]prop-2-enyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide
To be published
|
|
5FAL
 
 | | Crystal structure of PvHCT in complex with CoA and p-coumaroyl-shikimate | | Descriptor: | (3~{R},4~{R},5~{R})-5-[(~{E})-3-(4-hydroxyphenyl)prop-2-enoyl]oxy-3,4-bis(oxidanyl)cyclohexene-1-carboxylic acid, COENZYME A, GLYCEROL, ... | | Authors: | Pereira, J.H, Moriarty, N.W, Eudes, A, Yogiswara, S, Wang, G, Benites, V.T, Baidoo, E.E.K, Lee, T.S, Keasling, J.D, Loque, D, Adams, P.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin.
Plant Cell.Physiol., 57, 2016
|
|
5LQU
 
 | | Crystal Structure of COMT in complex with N-[(E)-3-[(2R,3S,4R,5R)-5-[6-(ethylamino)purin-9-yl]-3,4-dihydroxyoxolan-2-yl]prop-2-enyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Ellermann, M, Rudolph, M.G. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of COMT in complex with N-[(E)-3-[(2R,3S,4R,5R)-5-[6-(ethylamino)purin-9-yl]-3,4-dihydroxyoxolan-2-yl]prop-2-enyl]-5-(4-fluorophenyl)-2,3-dihydroxybenzamide
To be published
|
|
2JJ1
 
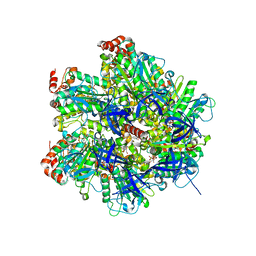 | | The Structure of F1-ATPase inhibited by piceatannol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
8HEK
 
 | |
