4YM7
 
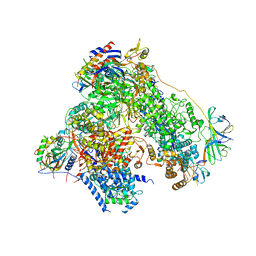 | | RNA polymerase I structure with an alternative dimer hinge | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Kostrewa, D, Kuhn, C.-D, Engel, C, Cramer, P. | | Deposit date: | 2015-03-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | An alternative RNA polymerase I structure reveals a dimer hinge.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8ISS
 
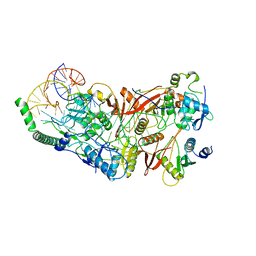 | | Cryo-EM structure of wild-type human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG at 3.19 A resolution | | Descriptor: | MAGNESIUM ION, RNA (88-MER), tRNA-splicing endonuclease subunit Sen15, ... | | Authors: | Sun, Y, Zhang, Y, Yuan, L, Han, Y. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Recognition and cleavage mechanism of intron-containing pre-tRNA by human TSEN endonuclease complex.
Nat Commun, 14, 2023
|
|
4YY3
 
 | | 30S ribosomal subunit- HigB complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schureck, M.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2015-03-23 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | mRNA bound to the 30S subunit is a HigB toxin substrate.
Rna, 22, 2016
|
|
2UXD
 
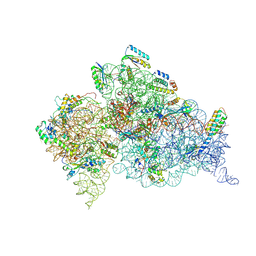 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
2UXB
 
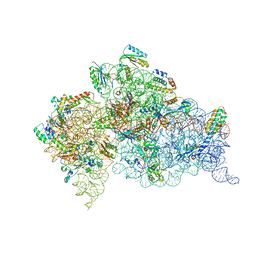 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT GGGU, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACCC, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit.
RNA, 13, 2007
|
|
7SCQ
 
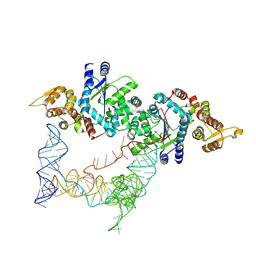 | |
7SC6
 
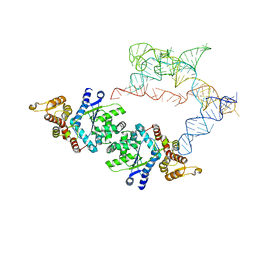 | |
7UXA
 
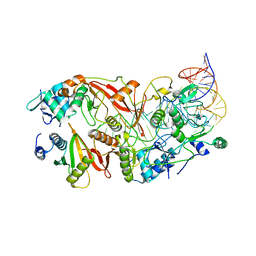 | | Human tRNA Splicing Endonuclease Complex bound to pre-tRNA-ARG | | Descriptor: | MAGNESIUM ION, RNA (78-MER), tRNA-splicing endonuclease subunit Sen15, ... | | Authors: | Stanley, R.E, Hayne, C.K. | | Deposit date: | 2022-05-05 | | Release date: | 2023-02-08 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for pre-tRNA recognition and processing by the human tRNA splicing endonuclease complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5ZAM
 
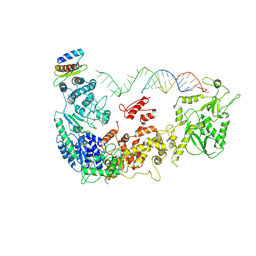 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
5ZAL
 
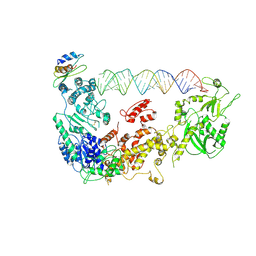 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2, RNA (73-mer) | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
7F1M
 
 | | Marburg virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Fujita, F.Y, Sugita, Y, Takamatsu, Y, Houri, K, Muramoto, Y, Nakano, M, Tsunoda, Y, Igarashi, M, Becker, S, Noda, T. | | Deposit date: | 2021-06-09 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Marburg virus nucleoprotein-RNA complex formation.
Nat Commun, 13, 2022
|
|
4QG3
 
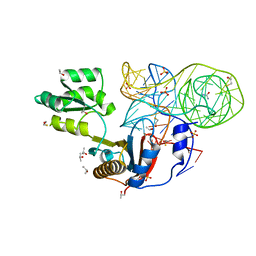 | | Crystal structure of mutant ribosomal protein G219V TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, BETA-MERCAPTOETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1C0A
 
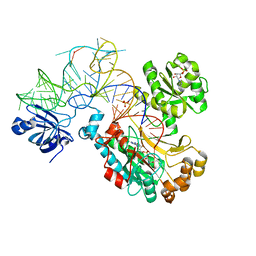 | | CRYSTAL STRUCTURE OF THE E. COLI ASPARTYL-TRNA SYNTHETASE : TRNAASP : ASPARTYL-ADENYLATE COMPLEX | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL TRNA, ASPARTYL TRNA SYNTHETASE, ... | | Authors: | Eiler, S, Dock-Bregeon, A.-C, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-07-15 | | Release date: | 1999-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis of aspartyl-tRNA(Asp) in Escherichia coli--a snapshot of the second step.
EMBO J., 18, 1999
|
|
6FQR
 
 | |
6GX6
 
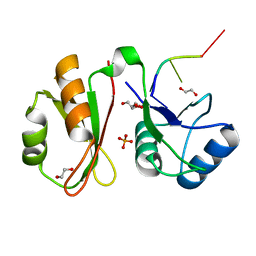 | | Crystal structure of IMP3 RRM12 in complex with RNA (ACAC) | | Descriptor: | 1,2-ETHANEDIOL, Insulin-like growth factor 2 mRNA-binding protein 3, PHOSPHATE ION, ... | | Authors: | Jia, M, Gut, H, Chao, A.J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of IMP3 RRM12 recognition of RNA.
RNA, 24, 2018
|
|
1VY5
 
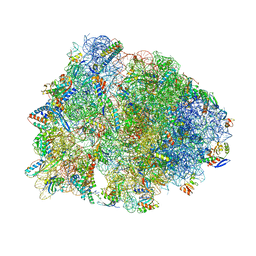 | | Crystal structure of the Thermus thermophilus 70S ribosome in the post-catalysis state of peptide bond formation containing dipeptydil-tRNA in the A site and deacylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6BY1
 
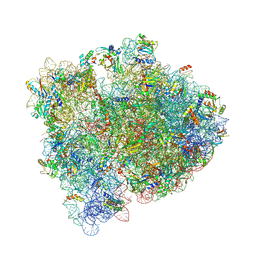 | | E. coli pH03H9 complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Amiri, H, Noller, H.F. | | Deposit date: | 2017-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Structural evidence for product stabilization by the ribosomal mRNA helicase.
Rna, 25, 2019
|
|
3SIU
 
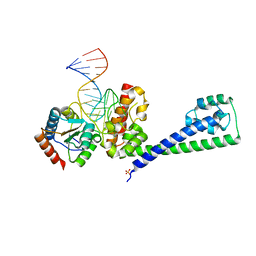 | | Structure of a hPrp31-15.5K-U4atac 5' stem loop complex, monomeric form | | Descriptor: | NHP2-like protein 1, SULFATE ION, U4/U6 small nuclear ribonucleoprotein Prp31, ... | | Authors: | Liu, S, Ghalei, H, Luhrmann, R, Wahl, M.C. | | Deposit date: | 2011-06-20 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | Structural basis for the dual U4 and U4atac snRNA-binding specificity of spliceosomal protein hPrp31.
Rna, 17, 2011
|
|
3CME
 
 | |
3CMA
 
 | |
5L4O
 
 | |
4QVI
 
 | | Crystal structure of mutant ribosomal protein M218L TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 50S ribosomal protein L1, ACETATE ION, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, NIkonov, S.V. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7KAB
 
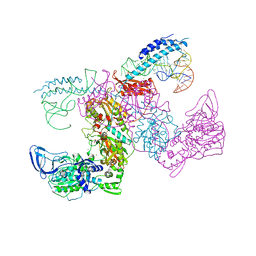 | | M. tuberculosis PheRS complex with cognate precursor tRNA and phenylalanine | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHENYLALANINE, ... | | Authors: | Chang, C, Michalska, K, Jedrzejczak, R, Wower, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
7K98
 
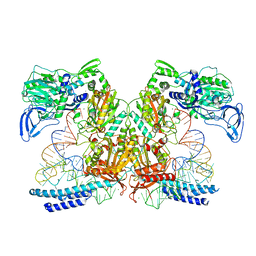 | | Preaminoacylation complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine (F-AMS) | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
7K9M
 
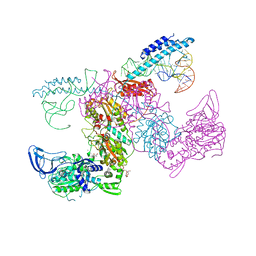 | | Crystal structure of the complex of M. tuberculosis PheRS with cognate precursor tRNA and 5'-O-(N-phenylalanyl)sulfamoyl-adenosine | | Descriptor: | 5'-O-(L-phenylalanylsulfamoyl)adenosine, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Michalska, K, Chang, C, Jedrzejczak, R, Wower, J, Baragana, B, Forte, B, Gilbert, I.H, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mycobacterium tuberculosis Phe-tRNA synthetase: structural insights into tRNA recognition and aminoacylation.
Nucleic Acids Res., 49, 2021
|
|
